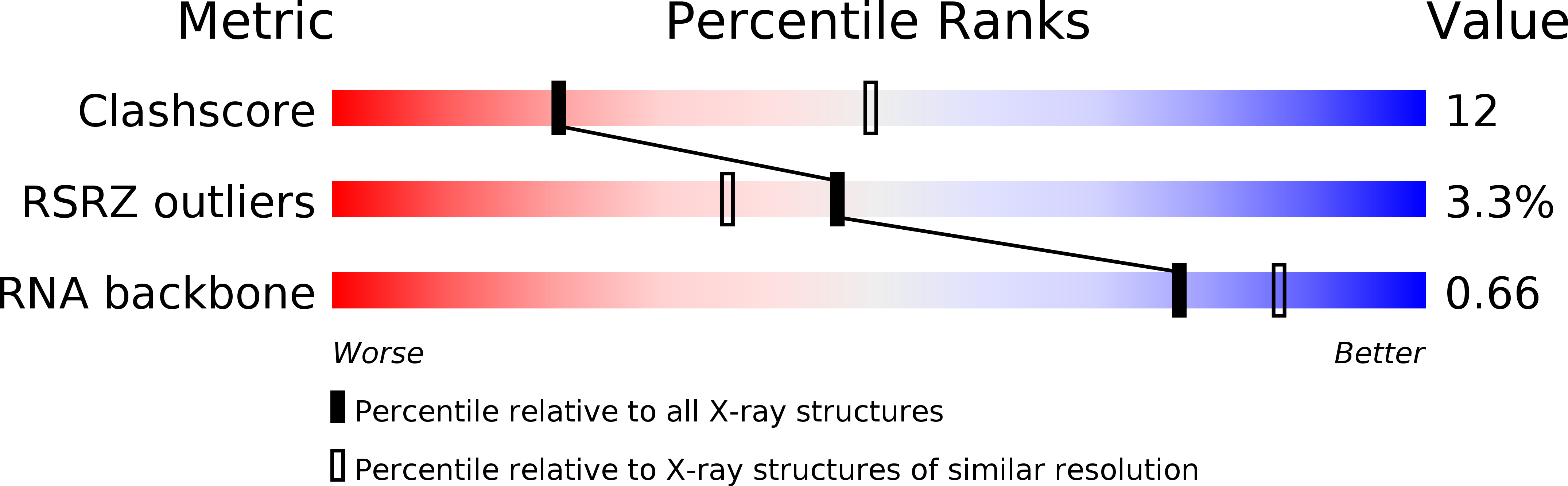Deposition Date
2009-06-29
Release Date
2009-11-03
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3I2U
Keywords:
Title:
Crystal structure of the haiprin ribozyme with a 2',5'-linked substrate and N1-deazaadenosine at position A10
Method Details:
Experimental Method:
Resolution:
2.80 Å
R-Value Free:
0.26
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 61 2 2