Deposition Date
2009-04-01
Release Date
2010-01-12
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3GX5
Keywords:
Title:
Crystal structure of T. tencongensis SAM-I riboswitch variant A94G/U34 bound with SAM
Method Details:
Experimental Method:
Resolution:
2.40 Å
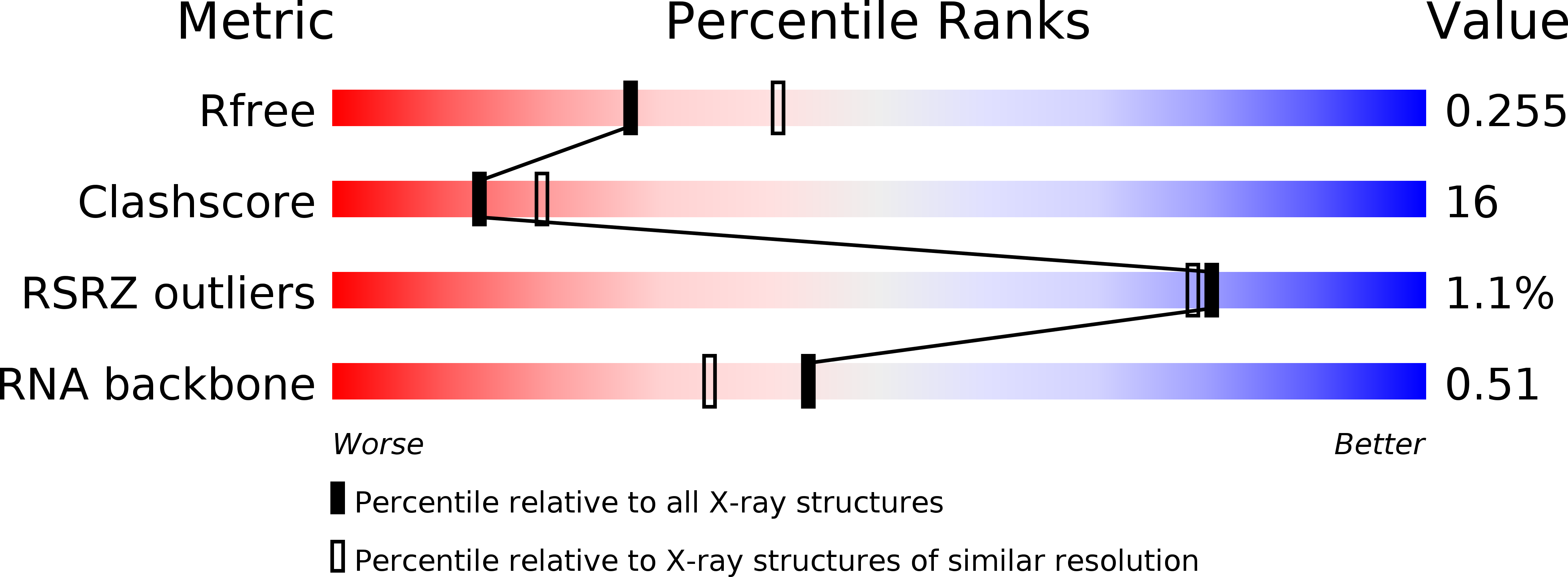
R-Value Free:
0.26
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 43 21 2