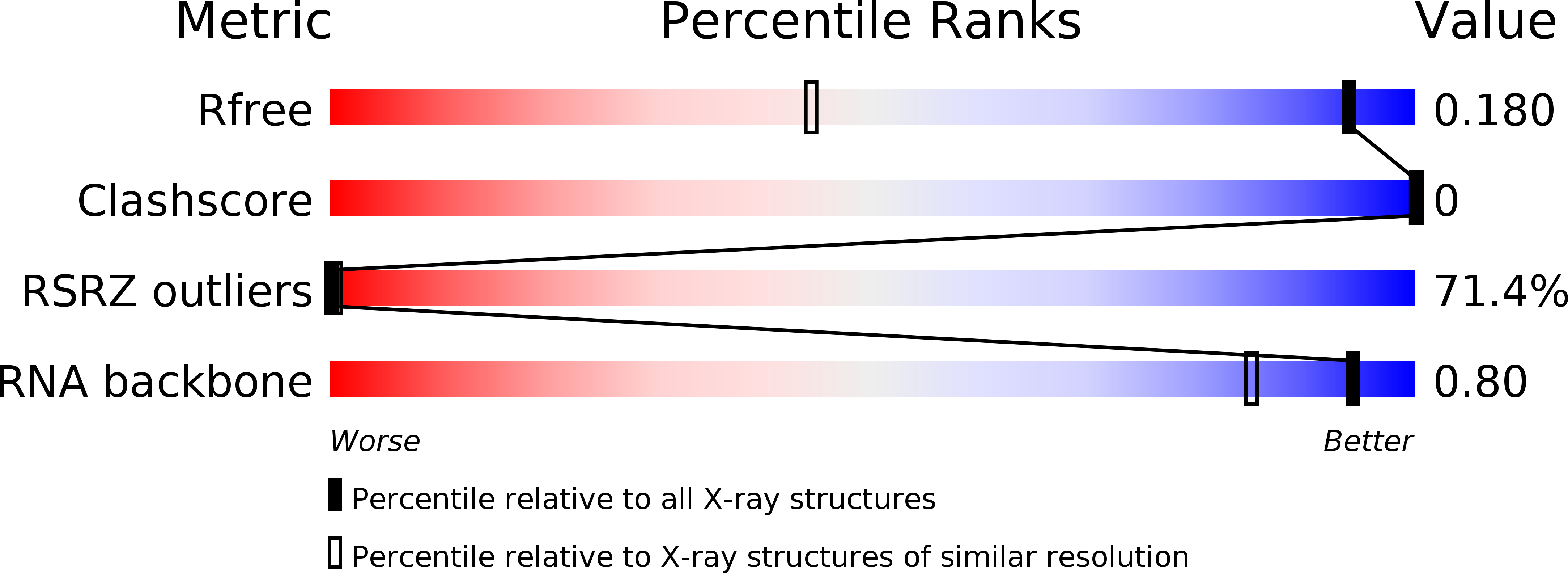Deposition Date
2009-03-31
Release Date
2009-07-28
Last Version Date
2023-11-01
Entry Detail
PDB ID:
3GVN
Keywords:
Title:
The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites
Method Details:
Experimental Method:
Resolution:
1.20 Å
R-Value Free:
0.20
R-Value Work:
0.19
Space Group:
C 1 2 1