Deposition Date
2009-02-04
Release Date
2009-12-01
Last Version Date
2023-09-06
Entry Detail
PDB ID:
3G4V
Keywords:
Title:
Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloropentane bound to the XE4 cavity
Biological Source:
Source Organism(s):
Scapharca inaequivalvis (Taxon ID: 6561)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.10 Å
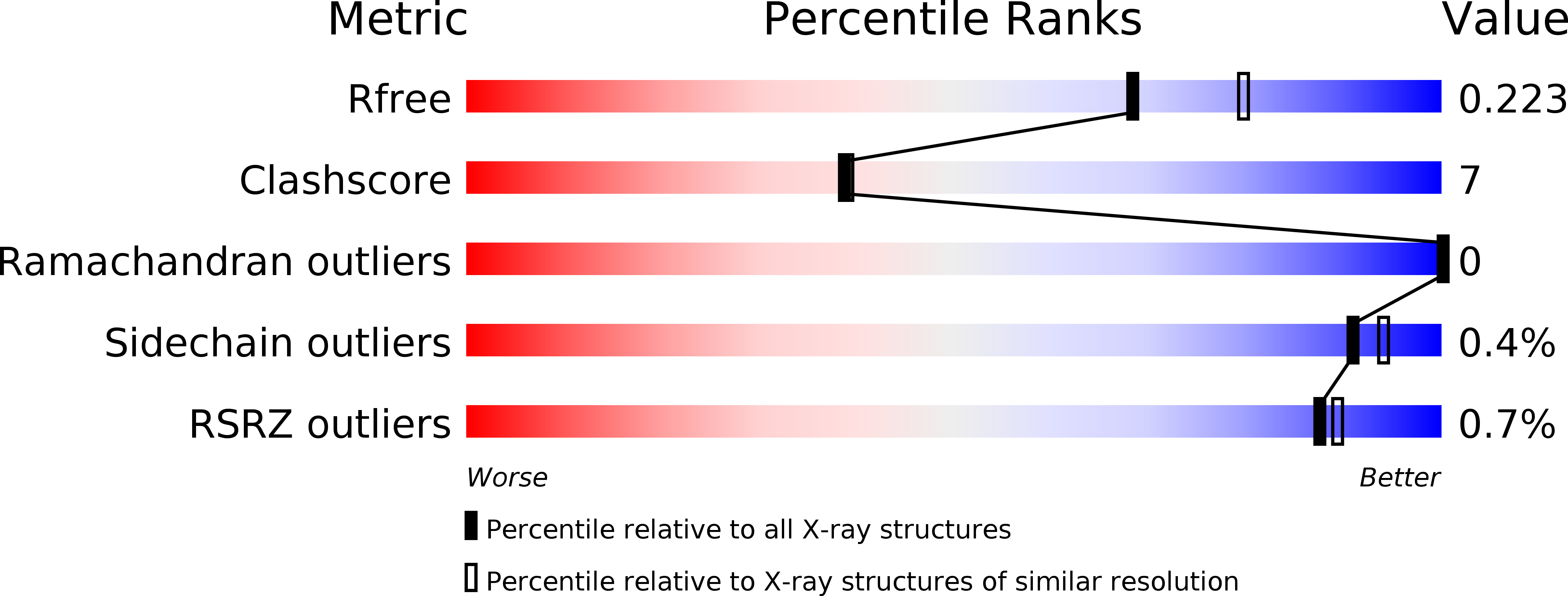
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
C 1 2 1