Deposition Date
2008-11-03
Release Date
2009-09-15
Last Version Date
2023-11-15
Entry Detail
PDB ID:
3F50
Keywords:
Title:
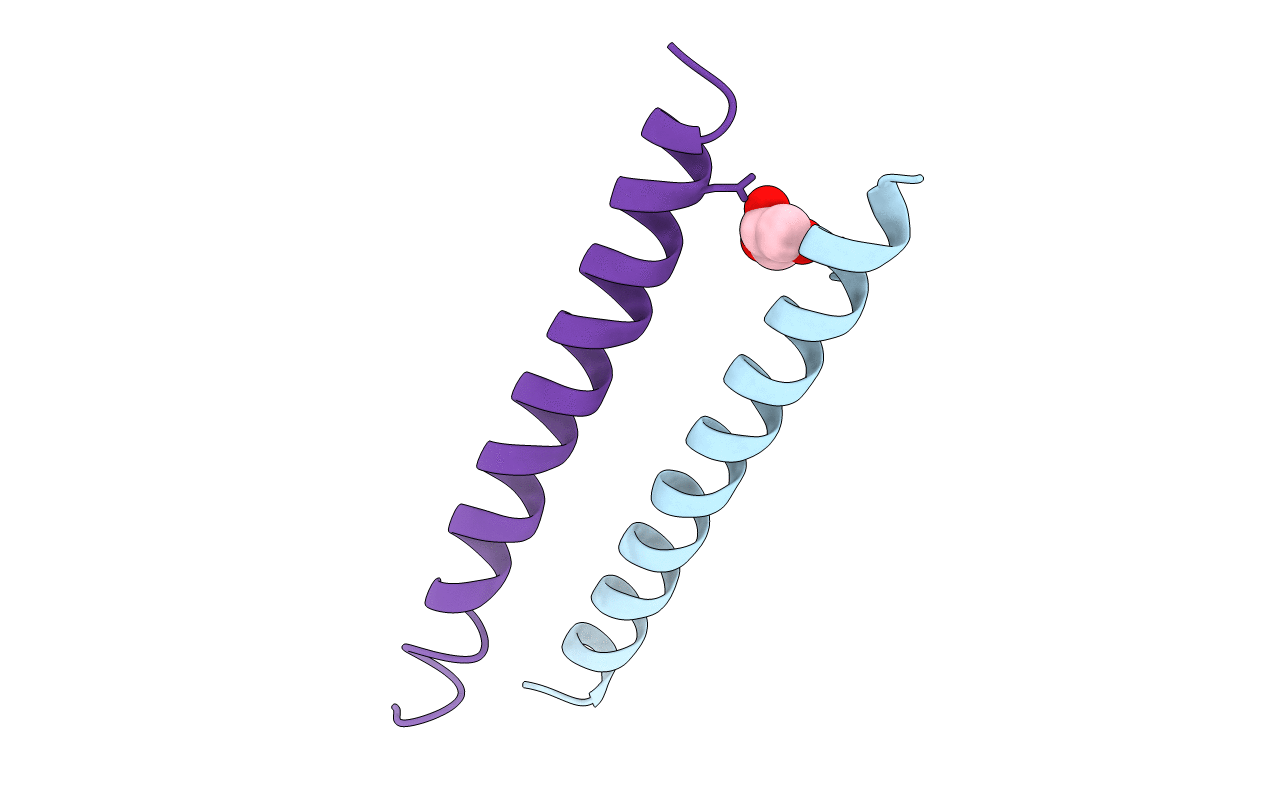
HIV gp41 six-helix bundle composed of an alpha/beta-peptide analogue of the CHR domain in complex with an NHR domain alpha-peptide
Method Details:
Experimental Method:
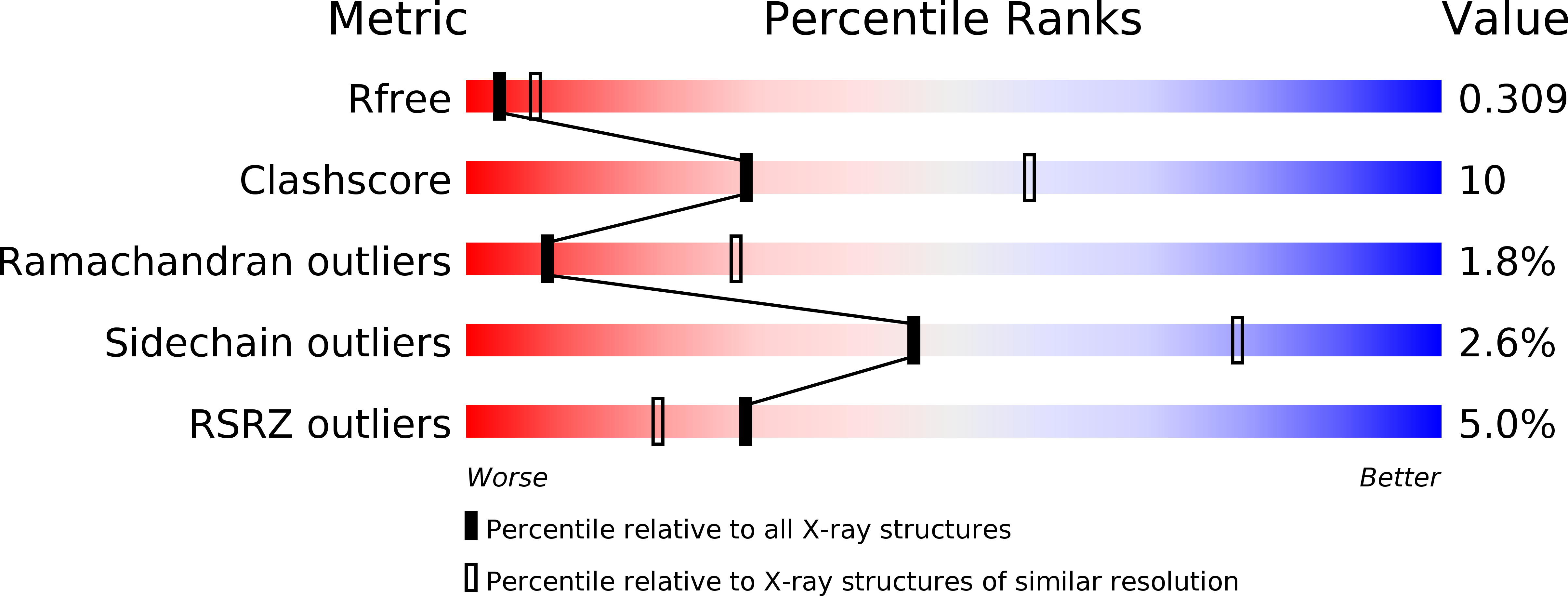
Resolution:
2.80 Å
R-Value Free:
0.30
R-Value Work:
0.26
R-Value Observed:
0.26
Space Group:
P 41 3 2