Deposition Date
2008-09-12
Release Date
2008-09-30
Last Version Date
2025-04-30
Entry Detail
PDB ID:
3EHB
Keywords:
Title:
A D-Pathway Mutation Decouples the Paracoccus Denitrificans Cytochrome c Oxidase by Altering the side chain orientation of a distant, conserved Glutamate
Biological Source:
Source Organism(s):
Paracoccus denitrificans (Taxon ID: 266)
Mus musculus (Taxon ID: 10090)
Mus musculus (Taxon ID: 10090)
Expression System(s):
Method Details:
Experimental Method:
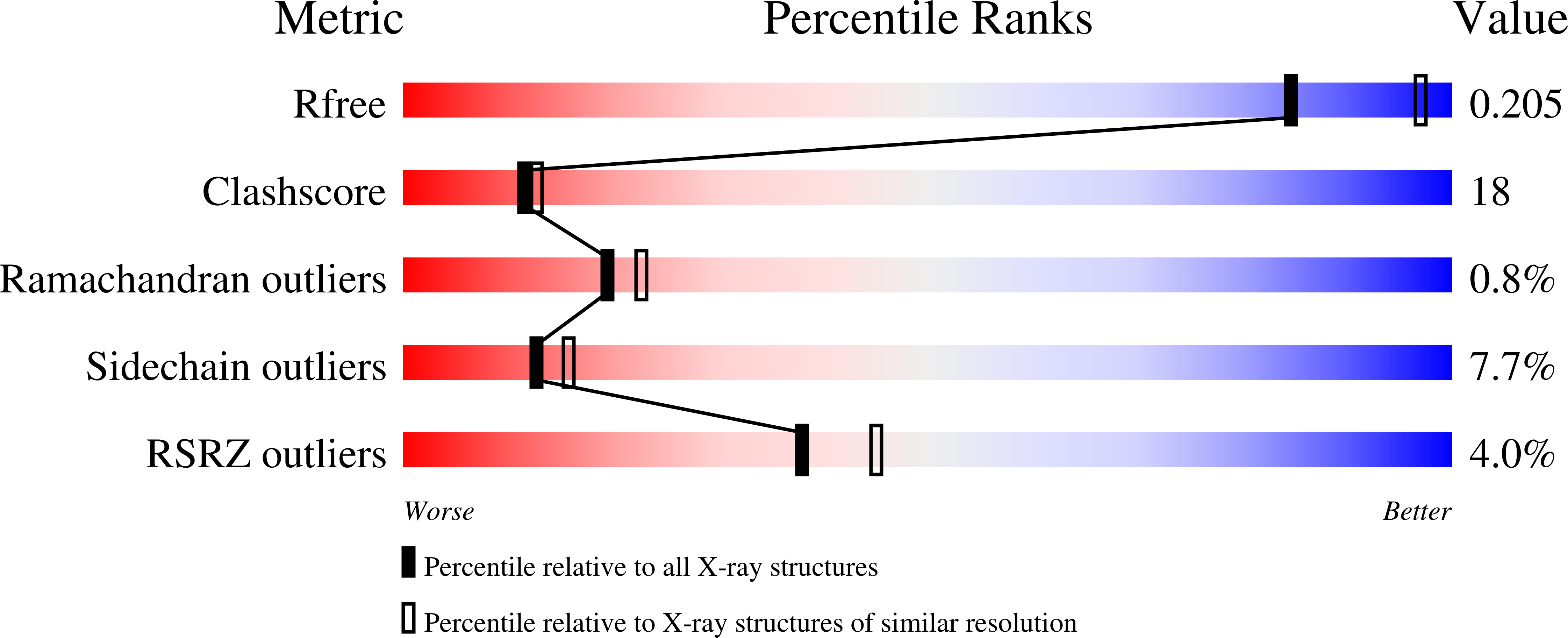
Resolution:
2.32 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21