Deposition Date
2008-09-03
Release Date
2008-10-28
Last Version Date
2023-11-15
Entry Detail
PDB ID:
3EDR
Keywords:
Title:
The crystal structure of caspase-7 in complex with Acetyl-LDESD-CHO
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Saccharomyces cerevisiae (Baker's yeast) (Taxon ID: 559292)
Saccharomyces cerevisiae (Baker's yeast) (Taxon ID: 559292)
Expression System(s):
Method Details:
Experimental Method:
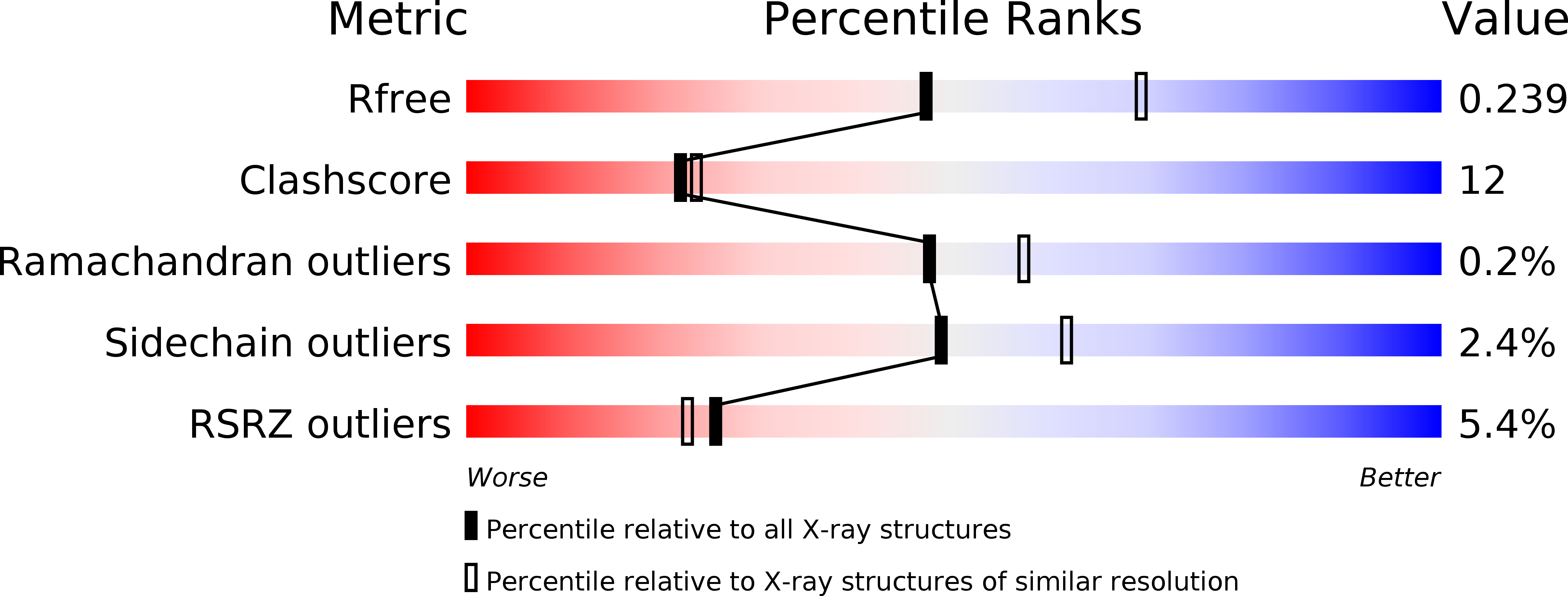
Resolution:
2.45 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.23
Space Group:
P 32 2 1