Deposition Date
2008-08-28
Release Date
2009-05-19
Last Version Date
2023-11-01
Entry Detail
PDB ID:
3EBN
Keywords:
Title:
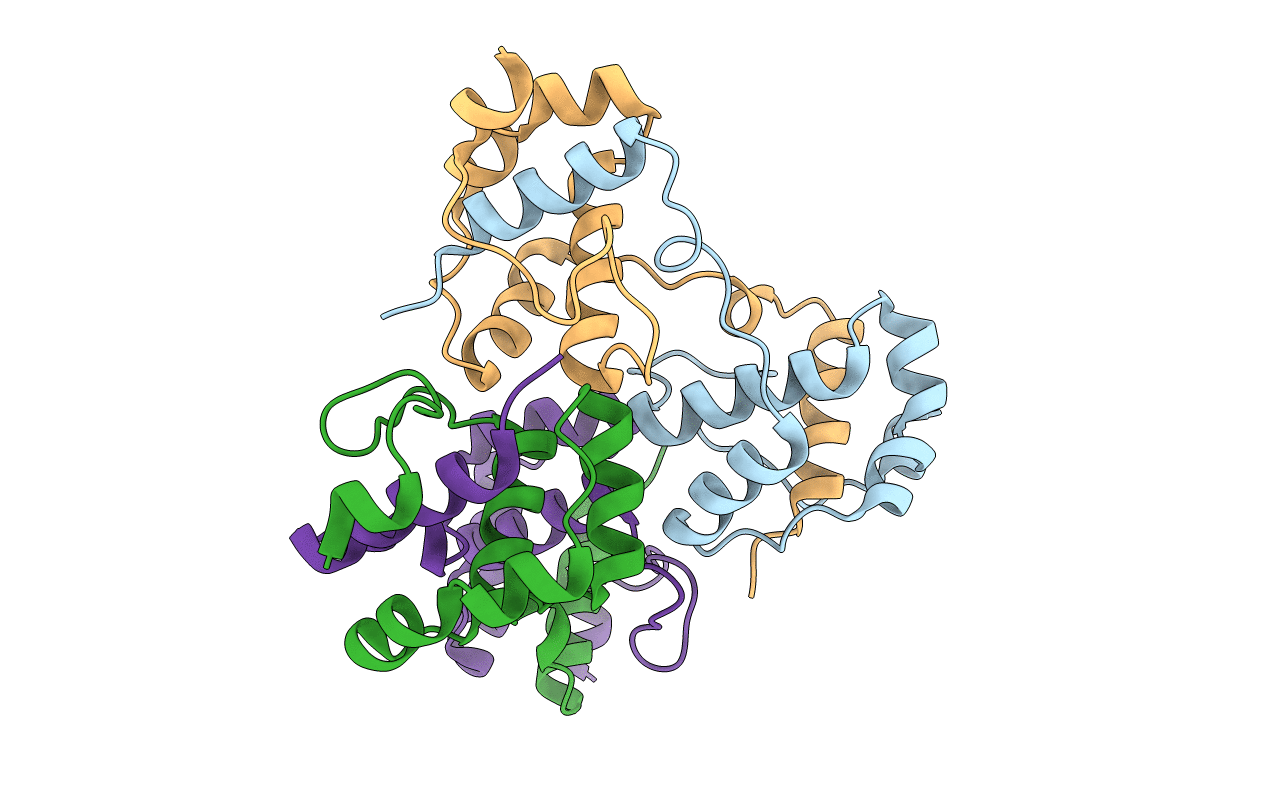
A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping
Biological Source:
Source Organism:
SARS coronavirus (Taxon ID: 227859)
Host Organism:
Method Details:
Experimental Method:
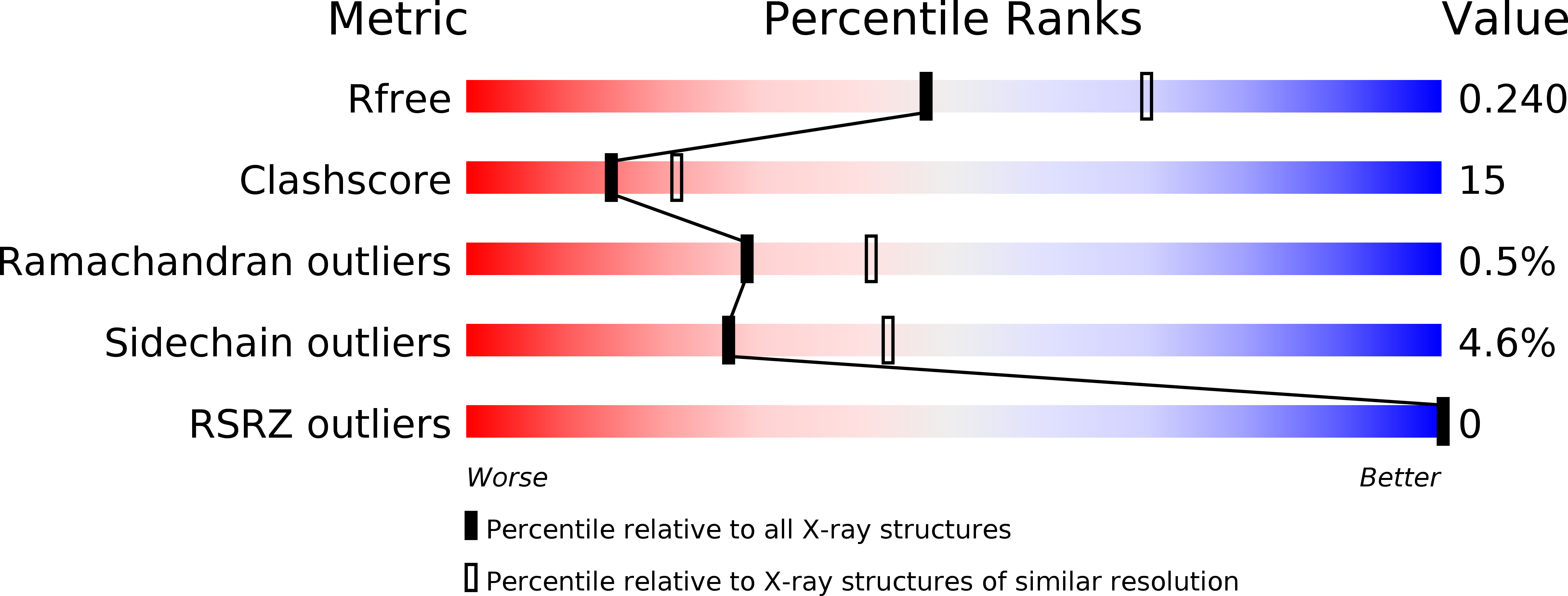
Resolution:
2.40 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1