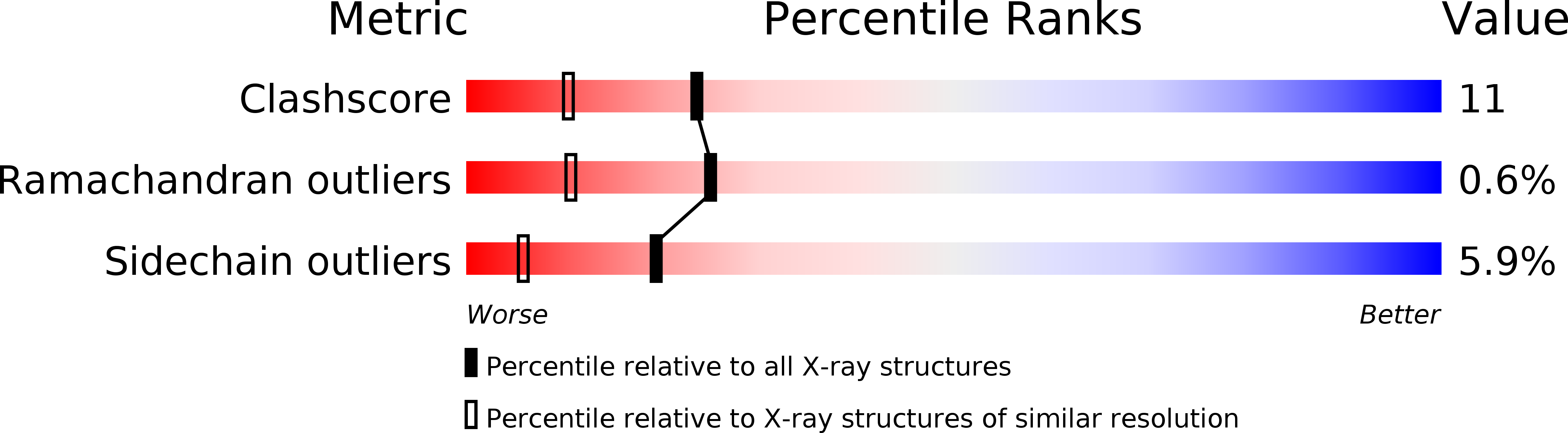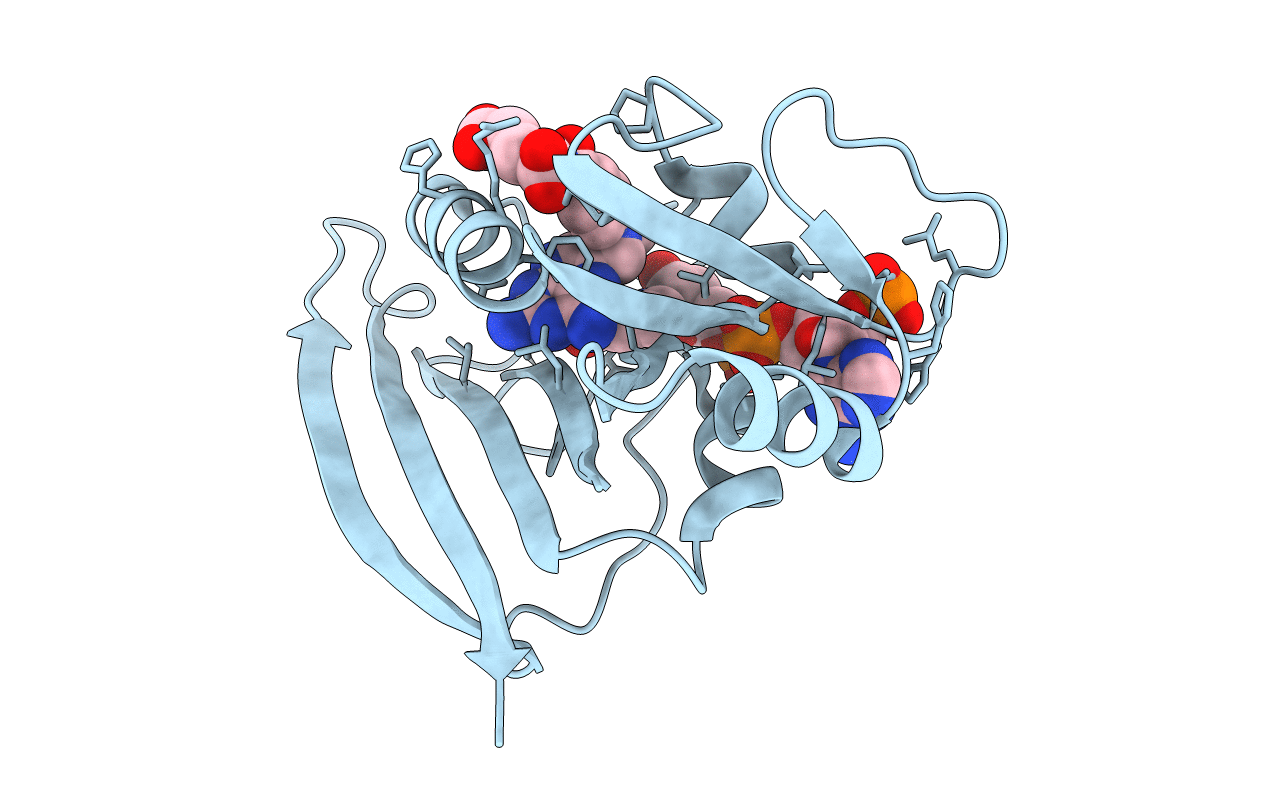
Deposition Date
1982-06-25
Release Date
1982-10-21
Last Version Date
2024-02-21
Entry Detail
PDB ID:
3DFR
Keywords:
Title:
CRYSTAL STRUCTURES OF ESCHERICHIA COLI AND LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE REFINED AT 1.7 ANGSTROMS RESOLUTION. I. GENERAL FEATURES AND BINDING OF METHOTREXATE
Biological Source:
Source Organism(s):
Lactobacillus casei (Taxon ID: 1582)
Method Details: