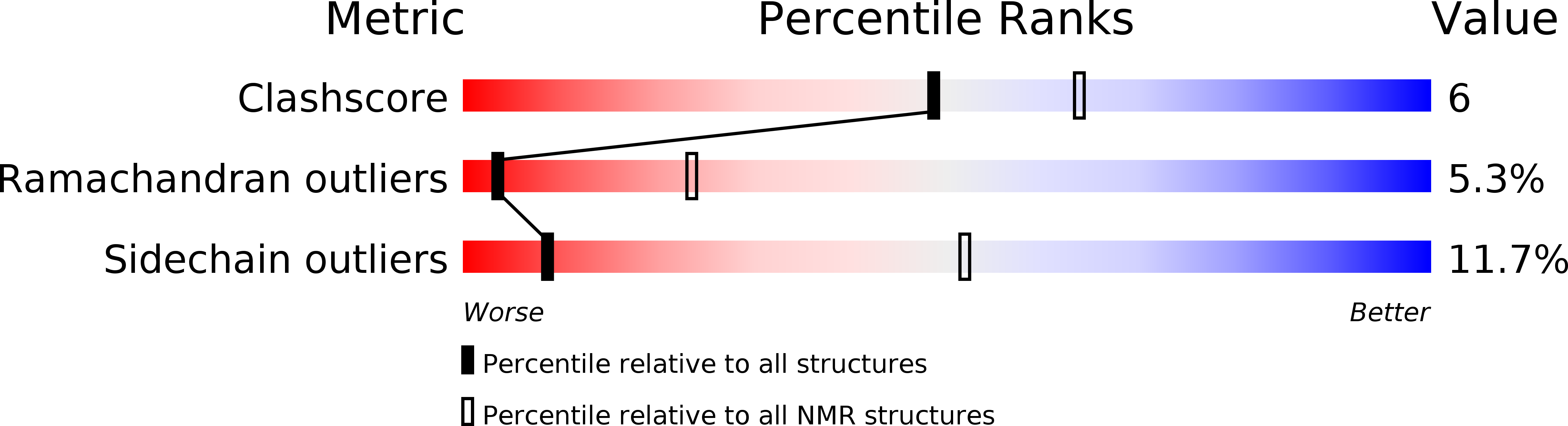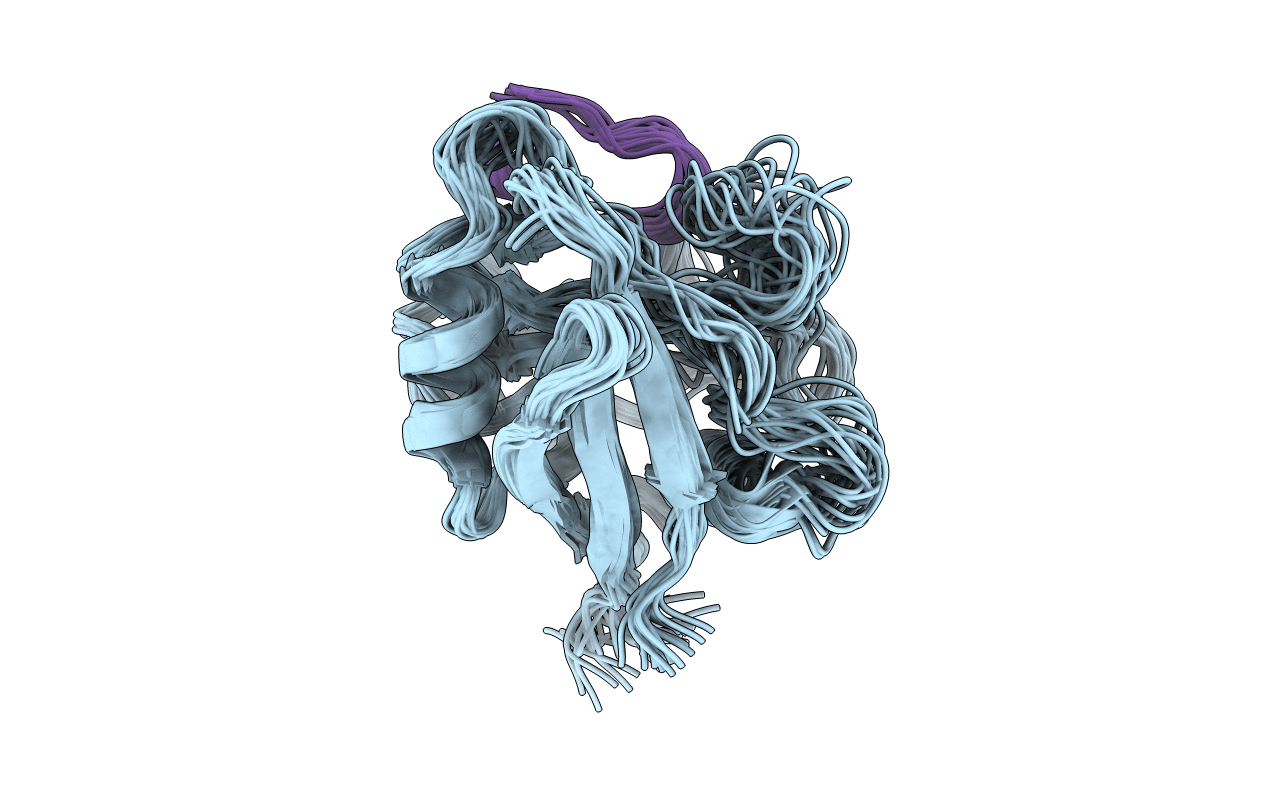
Deposition Date
1994-02-28
Release Date
1994-08-31
Last Version Date
2025-03-26
Entry Detail
PDB ID:
3CYS
Keywords:
Title:
DETERMINATION OF THE NMR SOLUTION STRUCTURE OF THE CYCLOPHILIN A-CYCLOSPORIN A COMPLEX
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
TOLYPOCLADIUM INFLATUM (Taxon ID: 29910)
TOLYPOCLADIUM INFLATUM (Taxon ID: 29910)
Method Details:
Experimental Method:
Conformers Submitted:
22