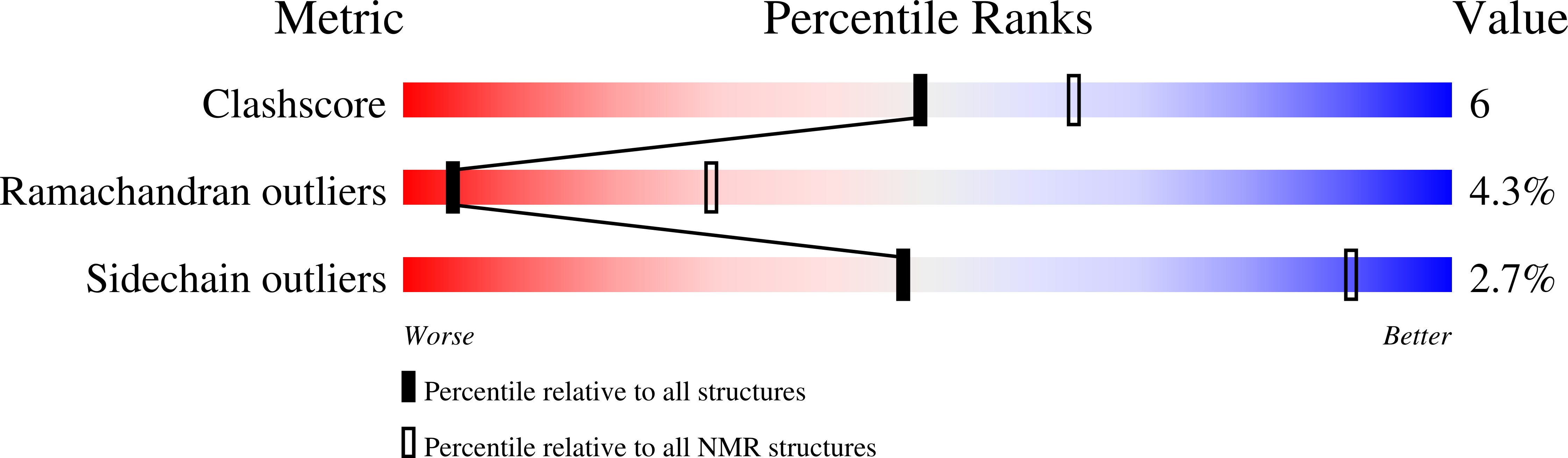
Deposition Date
1991-03-27
Release Date
1992-04-15
Last Version Date
2024-10-23
Entry Detail
PDB ID:
3CTI
Keywords:
Title:
RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF SQUASH TRYPSIN INHIBITOR
Biological Source:
Source Organism(s):
Cucurbita maxima (Taxon ID: 3661)
Method Details:
Experimental Method:
Conformers Submitted:
6