Deposition Date
2008-01-21
Release Date
2008-09-23
Last Version Date
2024-10-30
Entry Detail
PDB ID:
3C0J
Keywords:
Title:
Structure of E. coli dihydrodipicolinate synthase complexed with hydroxypyruvate
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.40 Å
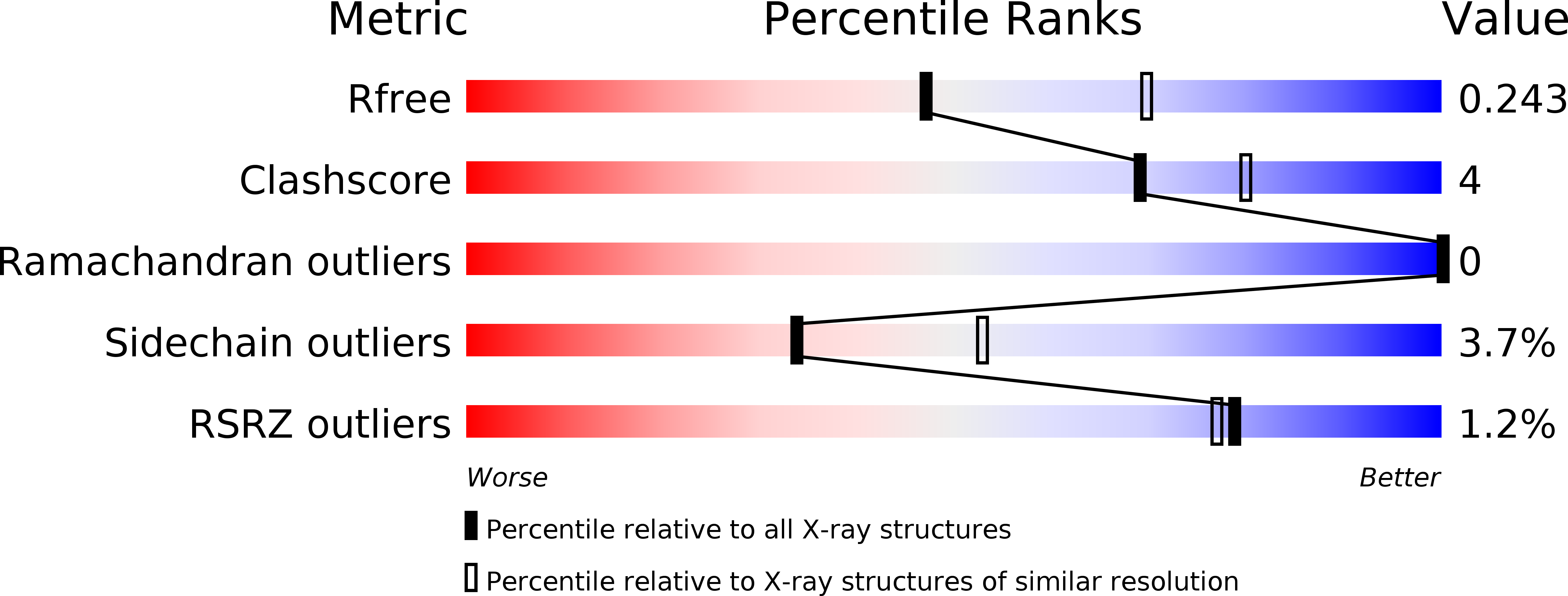
R-Value Free:
0.23
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 31 2 1