Deposition Date
2008-01-07
Release Date
2008-05-20
Last Version Date
2023-08-30
Entry Detail
PDB ID:
3BVD
Keywords:
Title:
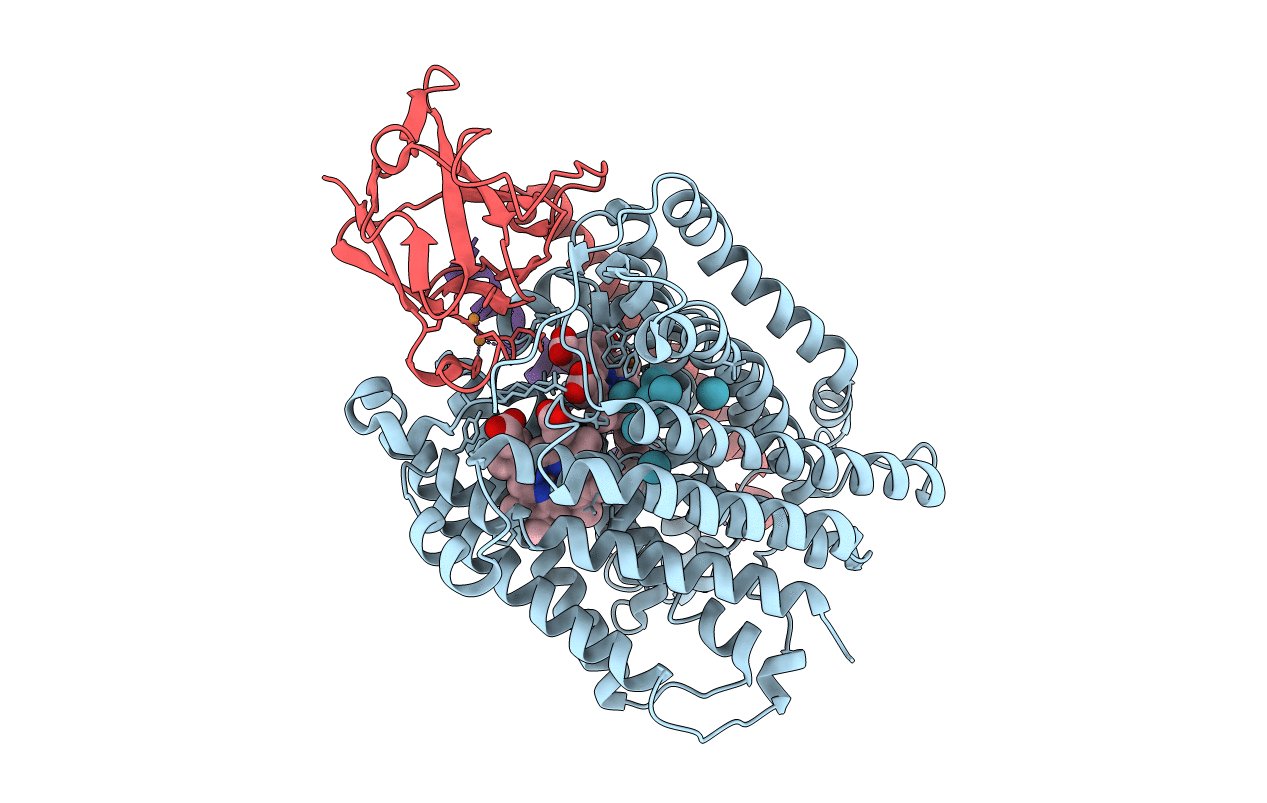
Structure of Surface-engineered Cytochrome ba3 Oxidase from Thermus thermophilus under Xenon Pressure, 100psi 5min
Biological Source:
Source Organism(s):
Thermus thermophilus (Taxon ID: )
Expression System(s):
Method Details:
Experimental Method:
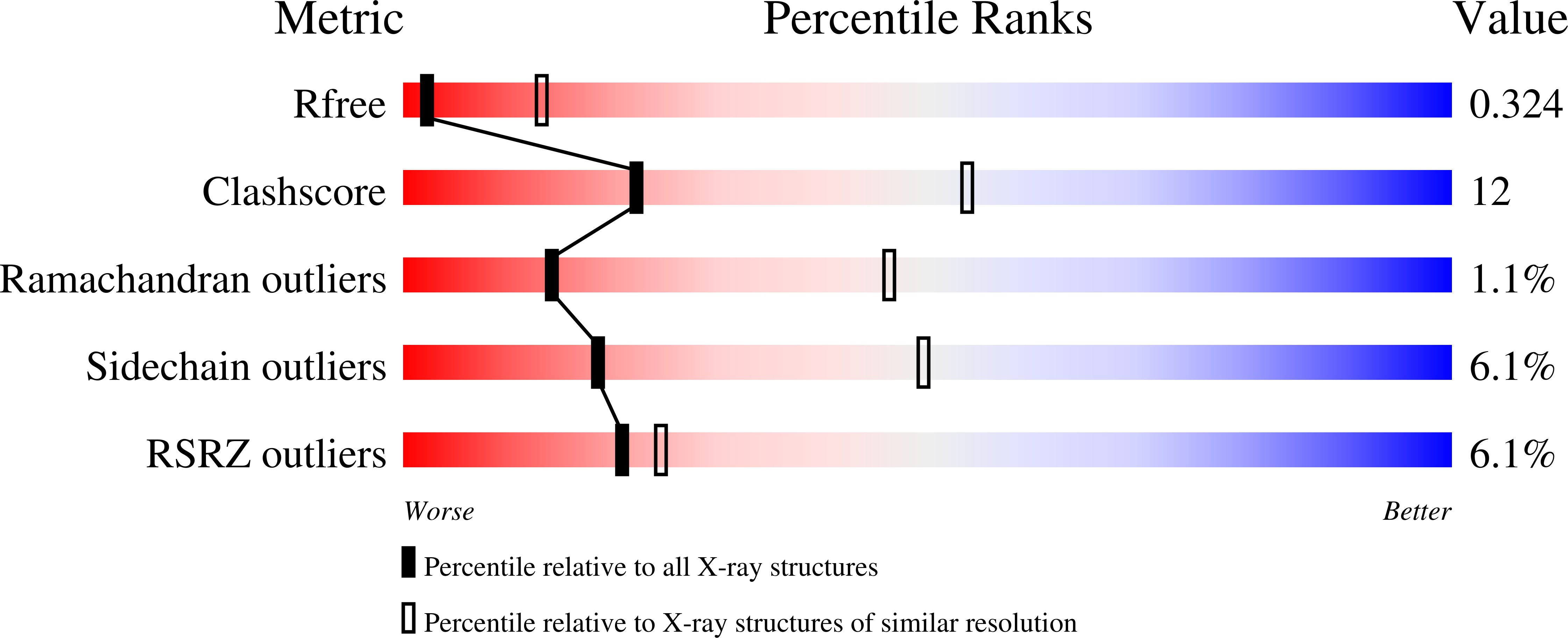
Resolution:
3.37 Å
R-Value Free:
0.33
R-Value Work:
0.29
Space Group:
P 41 21 2