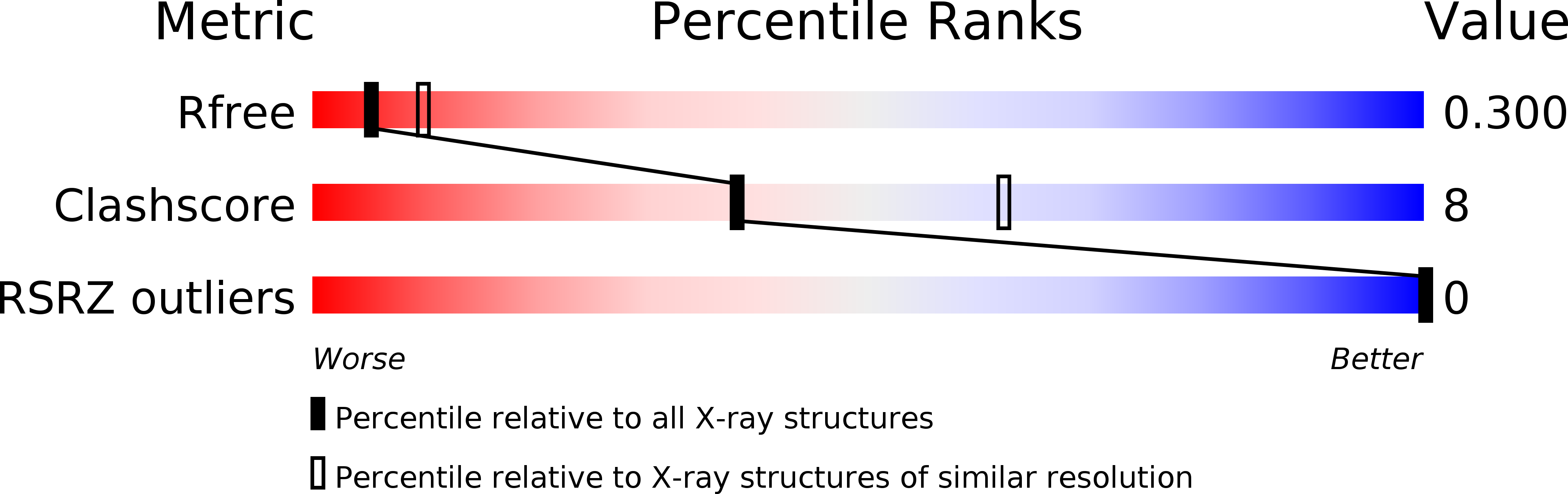Deposition Date
2010-06-07
Release Date
2011-04-27
Last Version Date
2023-11-01
Entry Detail
PDB ID:
3AJL
Keywords:
Title:
Crystal structure of d(CGCGGATf5UCGCG): 5-Formyluridine:guanosine Base-pair in B-DNA with DAPI
Method Details:
Experimental Method:
Resolution:
2.70 Å
R-Value Free:
0.28
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21