Deposition Date
2009-08-17
Release Date
2010-08-25
Last Version Date
2023-11-01
Entry Detail
PDB ID:
3A5Y
Keywords:
Title:
Crystal structure of GenX from Escherichia coli in complex with lysyladenylate analog
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
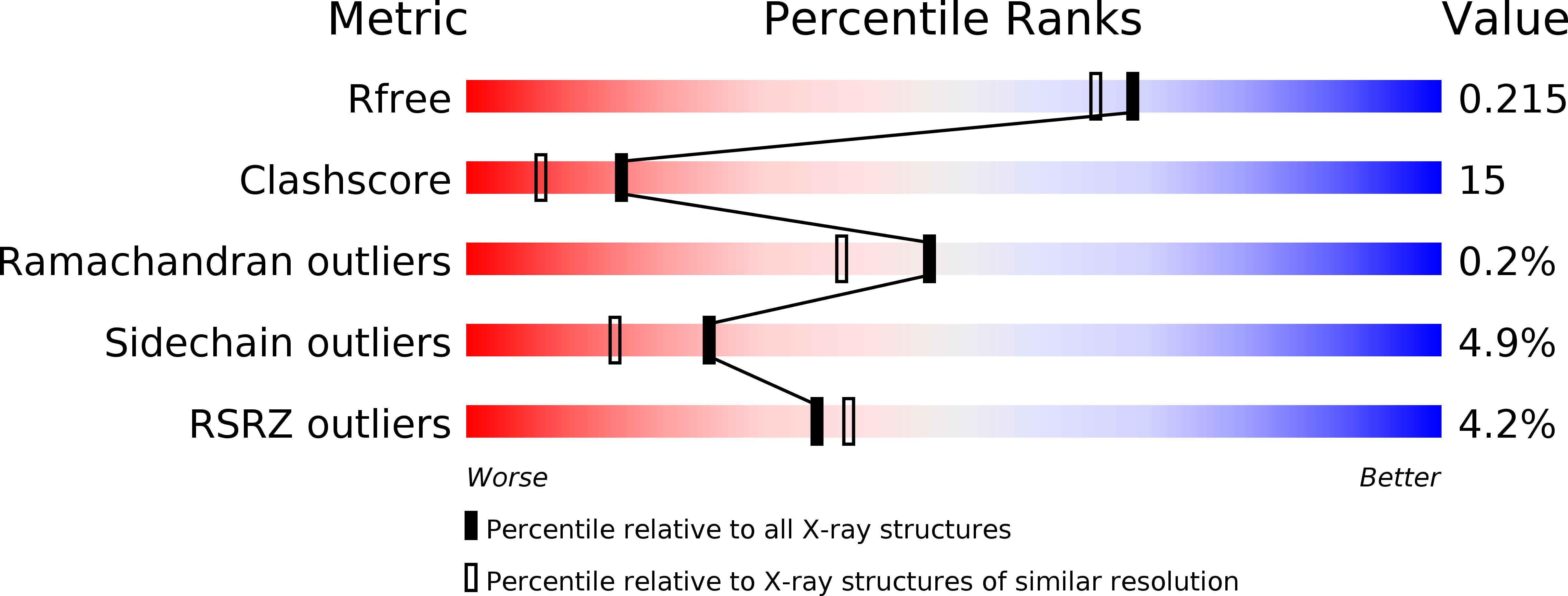
Resolution:
1.90 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1