Deposition Date
2009-06-23
Release Date
2010-01-12
Last Version Date
2023-11-15
Entry Detail
PDB ID:
3A3W
Keywords:
Title:
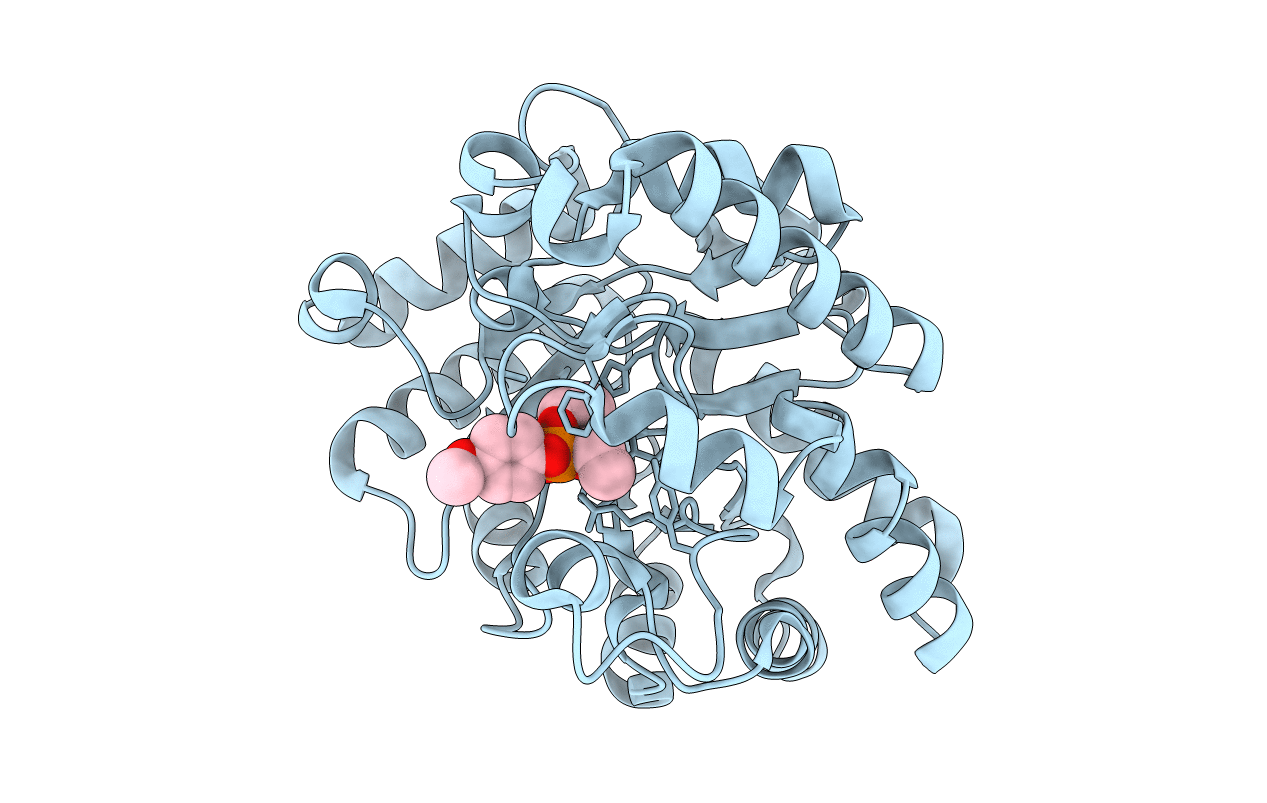
Structure of OpdA mutant (G60A/A80V/S92A/R118Q/K185R/Q206P/D208G/I260T/G273S) with diethyl 4-methoxyphenyl phosphate bound in the active site
Biological Source:
Source Organism(s):
Agrobacterium tumefaciens (Taxon ID: 358)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.85 Å
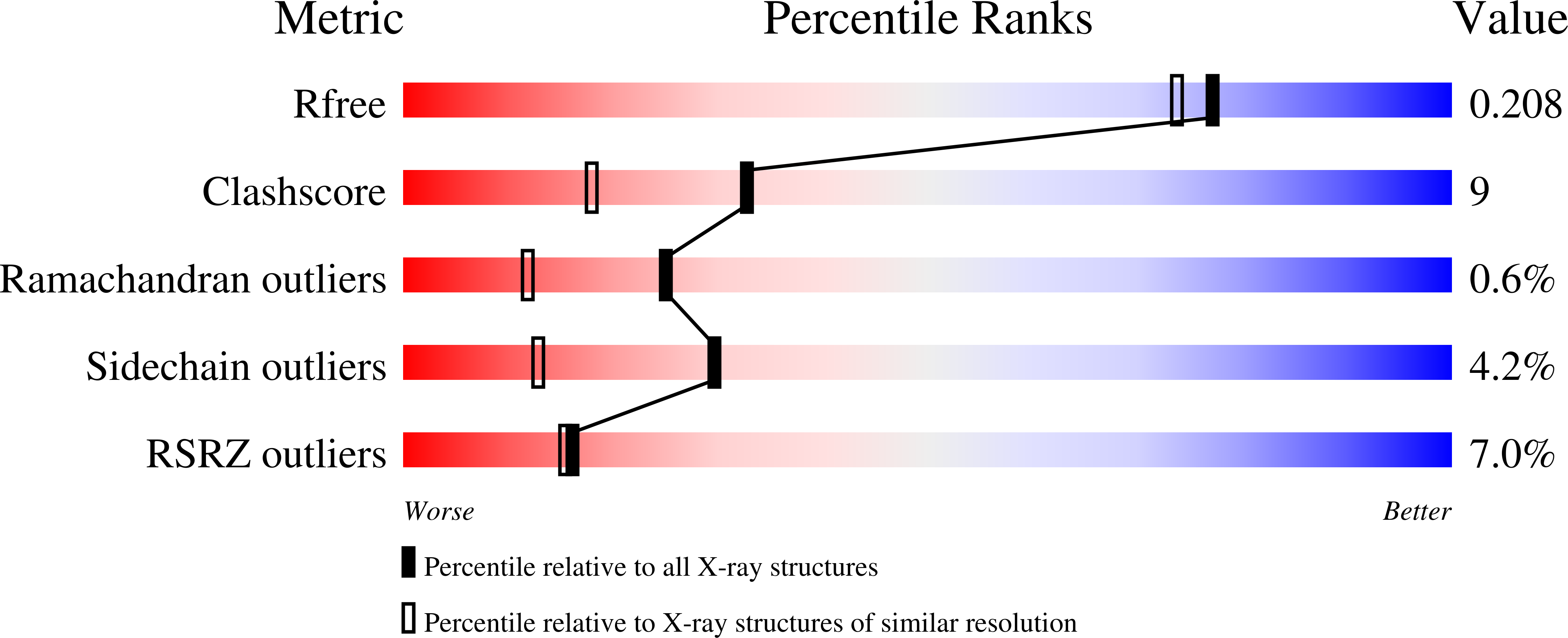
R-Value Free:
0.20
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 31 2 1