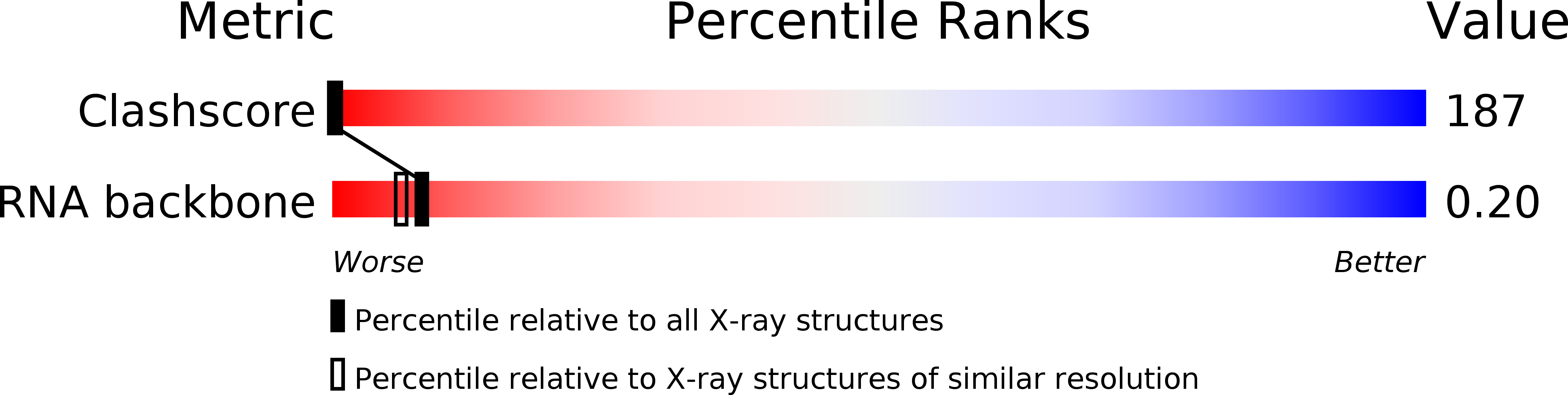
Deposition Date
1997-11-10
Release Date
1998-07-01
Last Version Date
2024-04-03
Entry Detail
PDB ID:
361D
Keywords:
Title:
CRYSTAL STRUCTURE OF DOMAIN E OF THERMUS FLAVUS 5S RRNA: A HELICAL RNA-STRUCTURE INCLUDING A TETRALOOP
Method Details:
Experimental Method:
Resolution:
3.00 Å
R-Value Free:
0.31
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 32 2 1