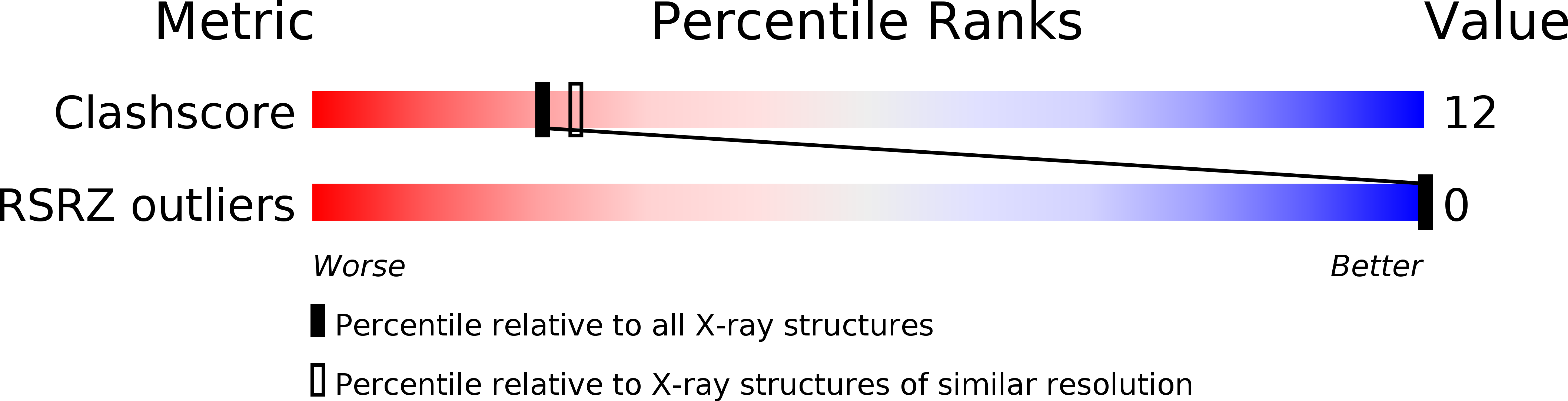
Deposition Date
1996-06-26
Release Date
1997-01-20
Last Version Date
2024-04-03
Entry Detail
PDB ID:
303D
Keywords:
Title:
META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2
Method Details:
Experimental Method:
Resolution:
2.20 Å
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21