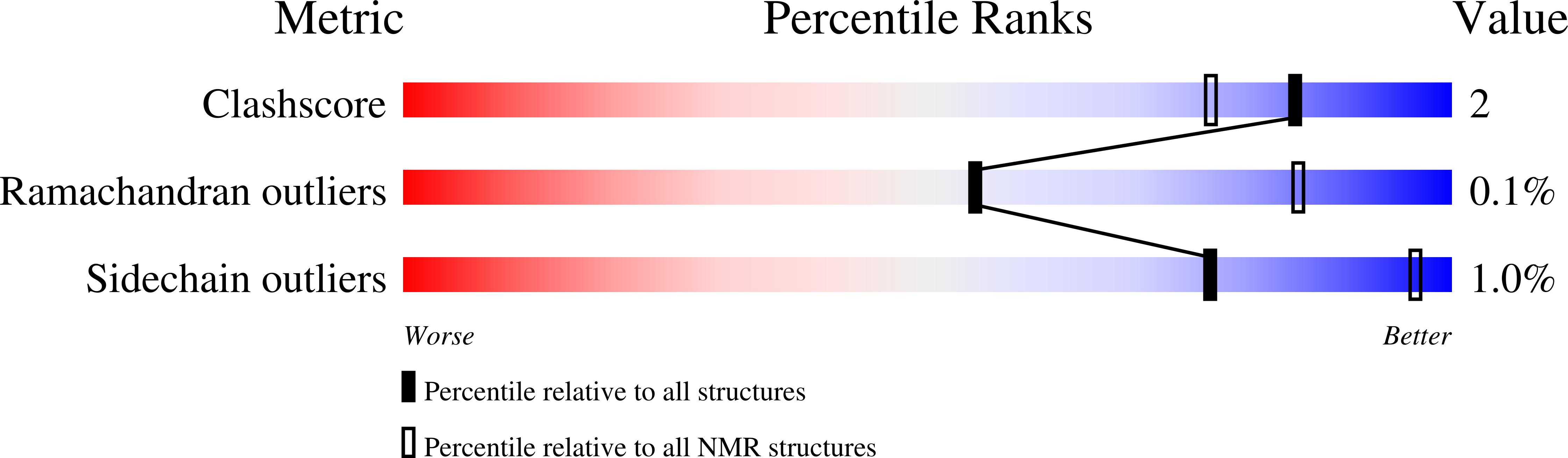
Deposition Date
2009-05-03
Release Date
2009-06-30
Last Version Date
2024-05-22
Entry Detail
PDB ID:
2KIF
Keywords:
Title:
Solution NMR structure of an O6-methylguanine DNA methyltransferase family protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium target VpR247.
Biological Source:
Source Organism(s):
Vibrio parahaemolyticus AQ3810 (Taxon ID: 419109)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy