Deposition Date
2012-11-05
Release Date
2012-11-21
Last Version Date
2023-12-20
Entry Detail
PDB ID:
2YQ5
Keywords:
Title:
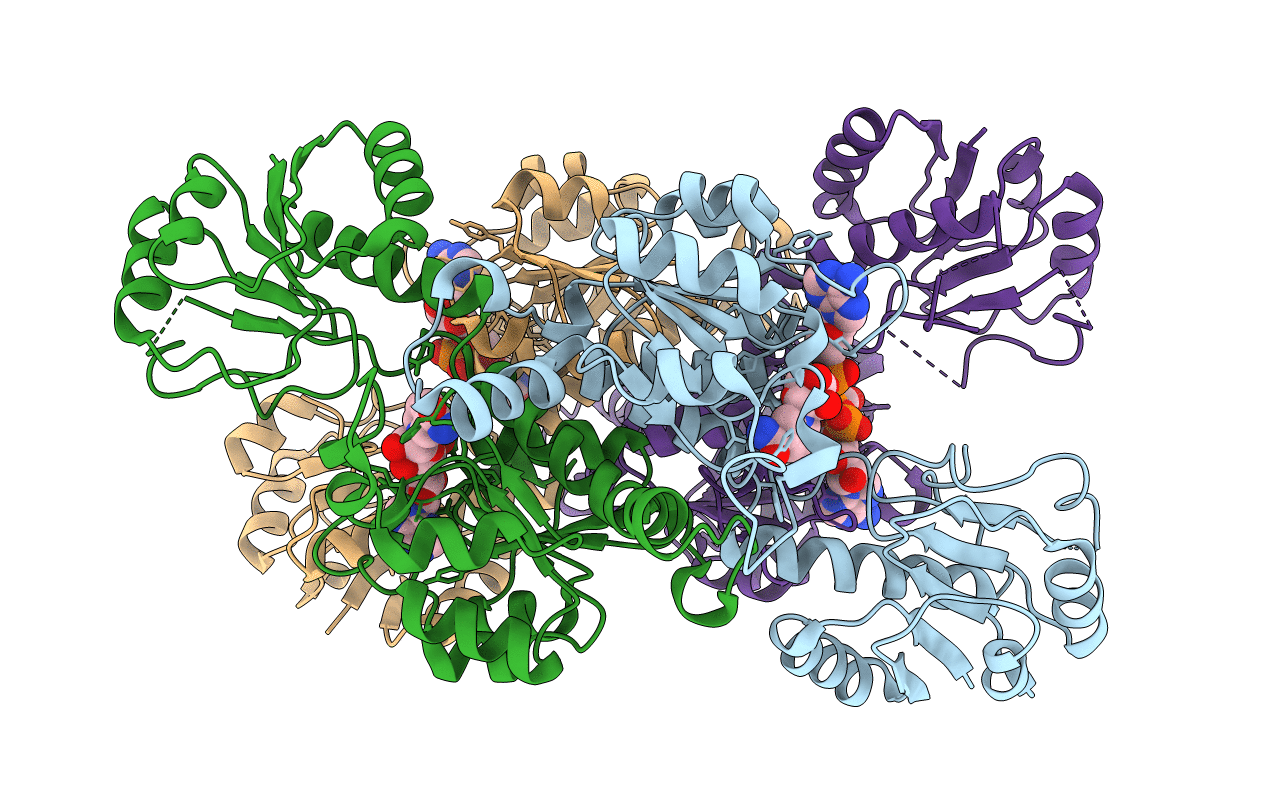
Crystal Structure of D-isomer specific 2-hydroxyacid dehydrogenase from Lactobacillus delbrueckii ssp. bulgaricus: NAD complexed form
Biological Source:
Source Organism(s):
LACTOBACILLUS DELBRUECKII SUBSP. BULGARICUS (Taxon ID: 1585)
Expression System(s):
Method Details:
Experimental Method:
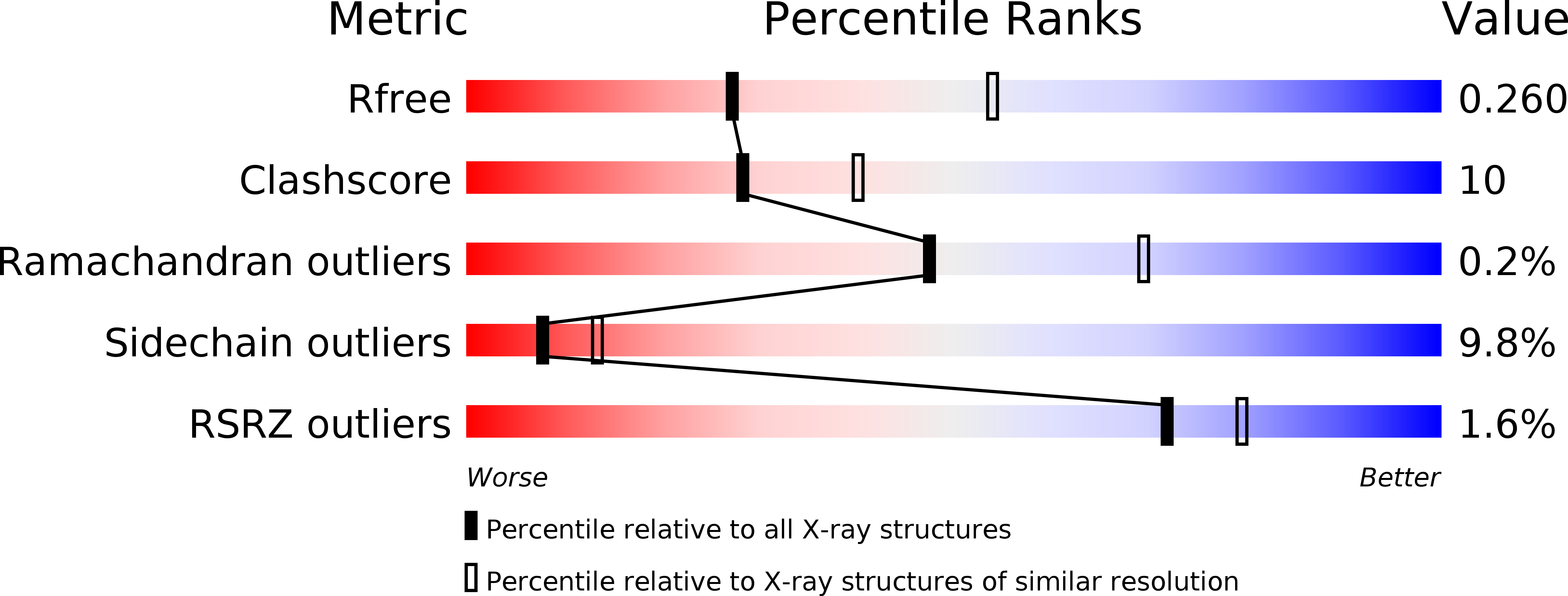
Resolution:
2.75 Å
R-Value Free:
0.25
R-Value Work:
0.18
R-Value Observed:
0.19
Space Group:
P 21 21 21