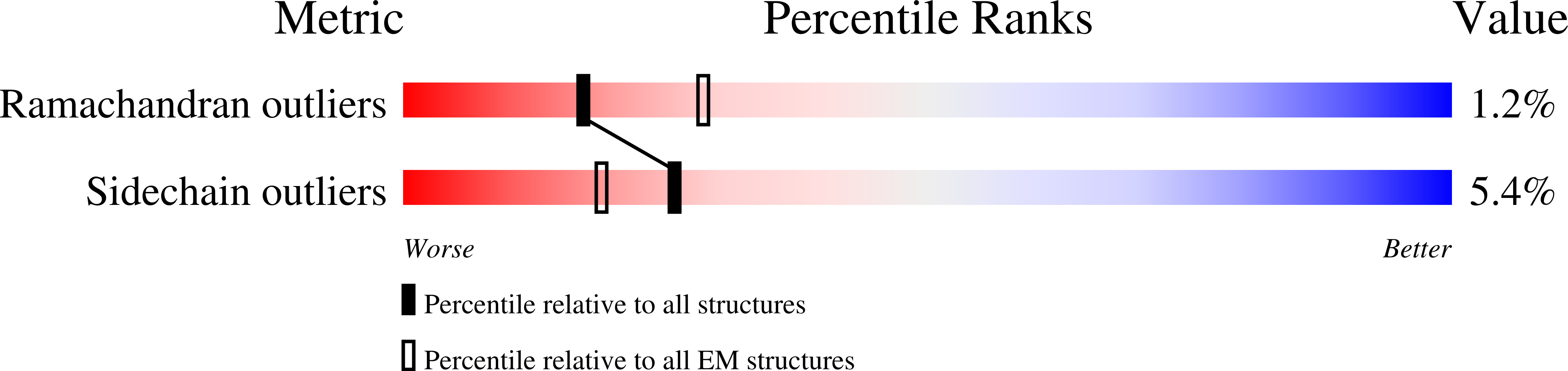
Deposition Date
2011-03-02
Release Date
2011-10-19
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2YBB
Keywords:
Title:
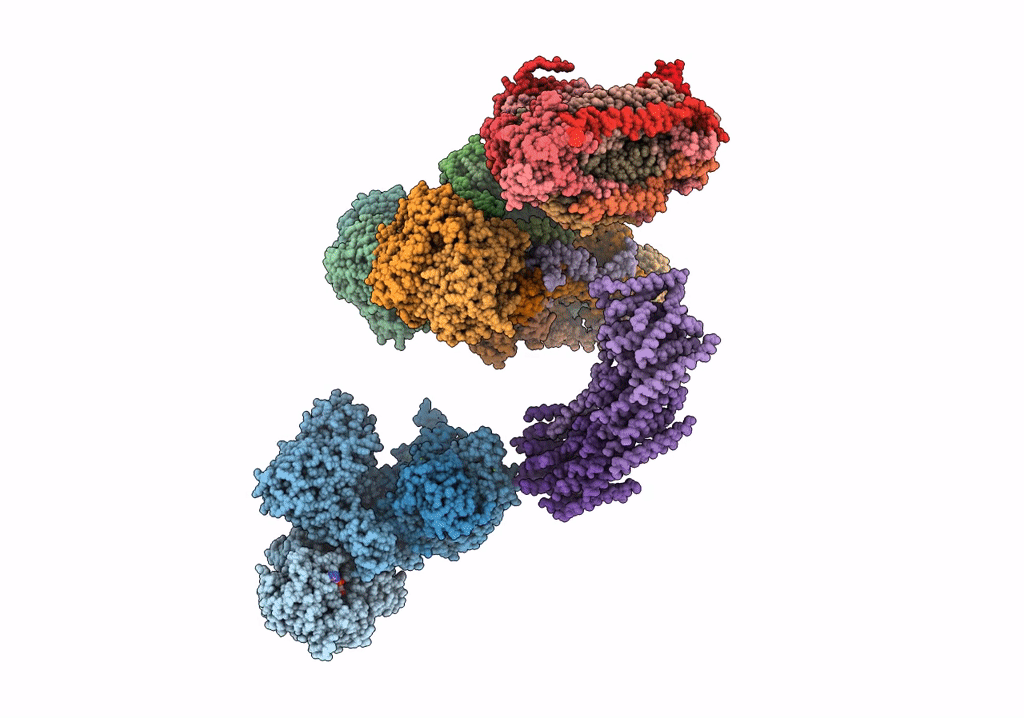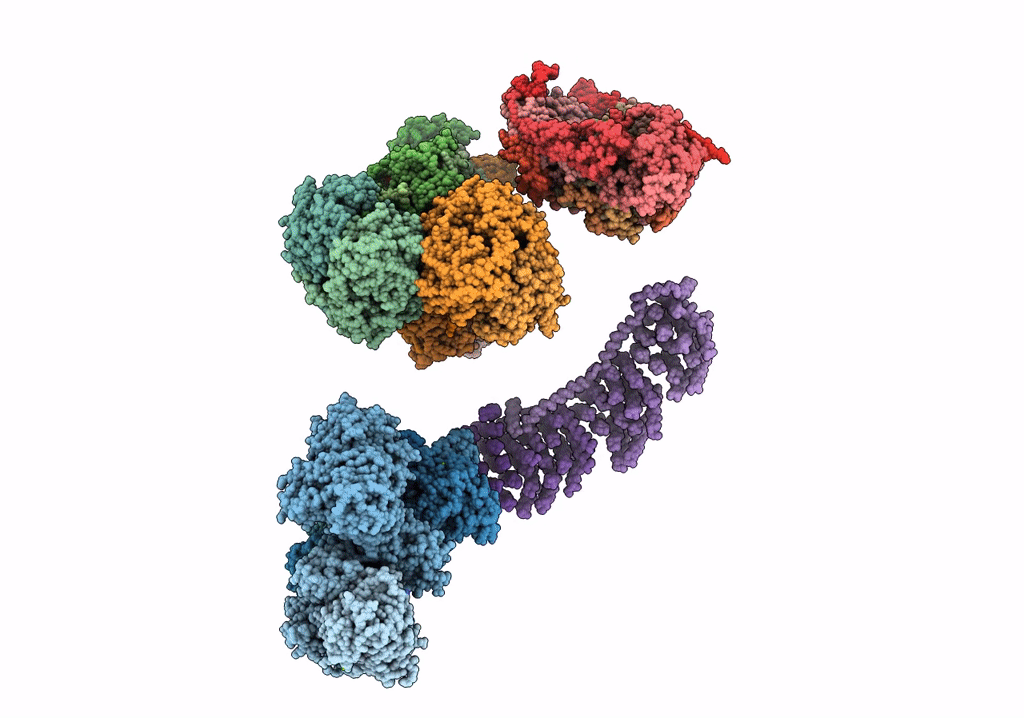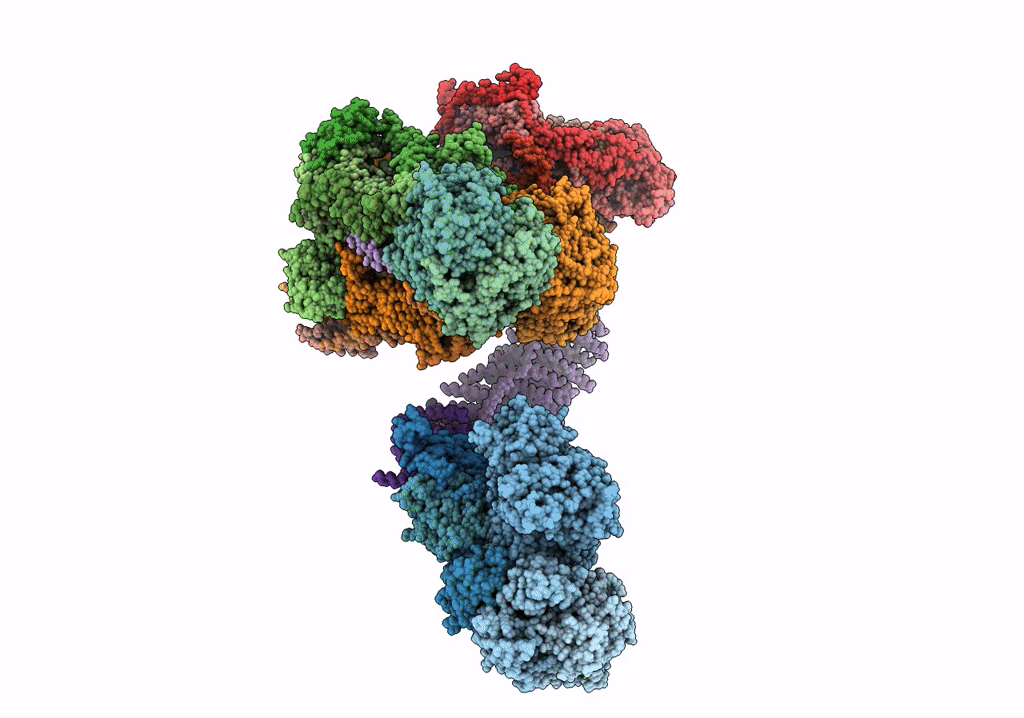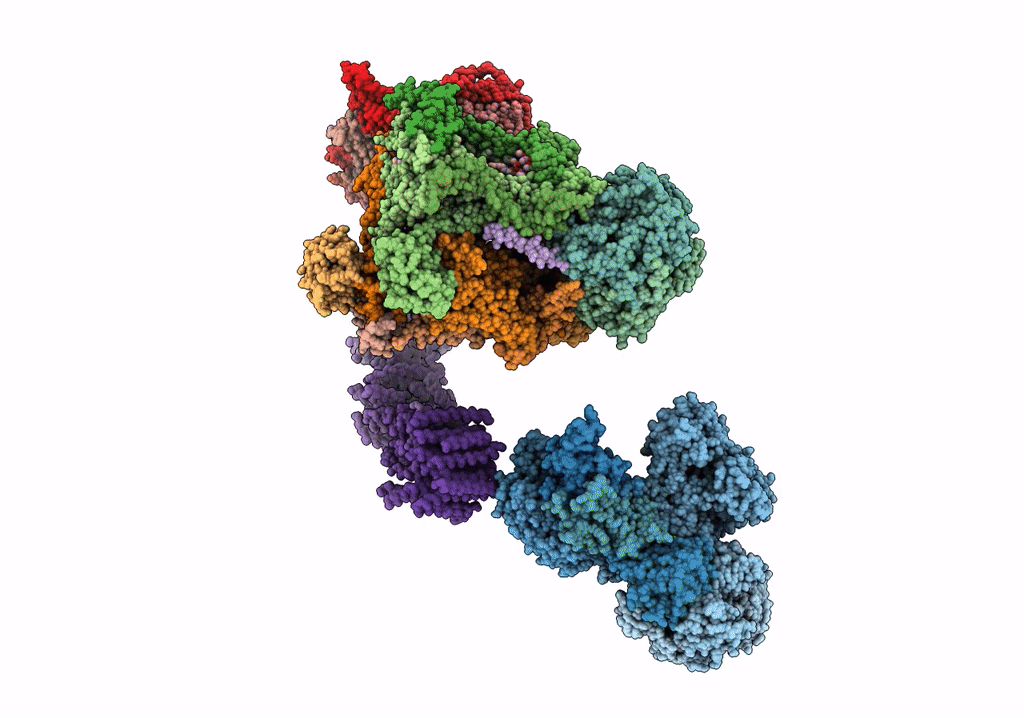
Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876)
Biological Source:
Source Organism(s):
THERMUS THERMOPHILUS (Taxon ID: 300852)
BOS TAURUS (Taxon ID: 9913)
ESCHERICHIA COLI (Taxon ID: 511693)
BOS TAURUS (Taxon ID: 9913)
ESCHERICHIA COLI (Taxon ID: 511693)
Method Details:
Experimental Method:
Resolution:
19.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE