Deposition Date
2011-01-28
Release Date
2011-06-15
Last Version Date
2023-12-20
Entry Detail
PDB ID:
2Y74
Keywords:
Title:
THE CRYSTAL STRUCTURE OF HUMAN SOLUBLE PRIMARY AMINE OXIDASE AOC3 IN THE OFF-COPPER CONFORMATION
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
Method Details:
Experimental Method:
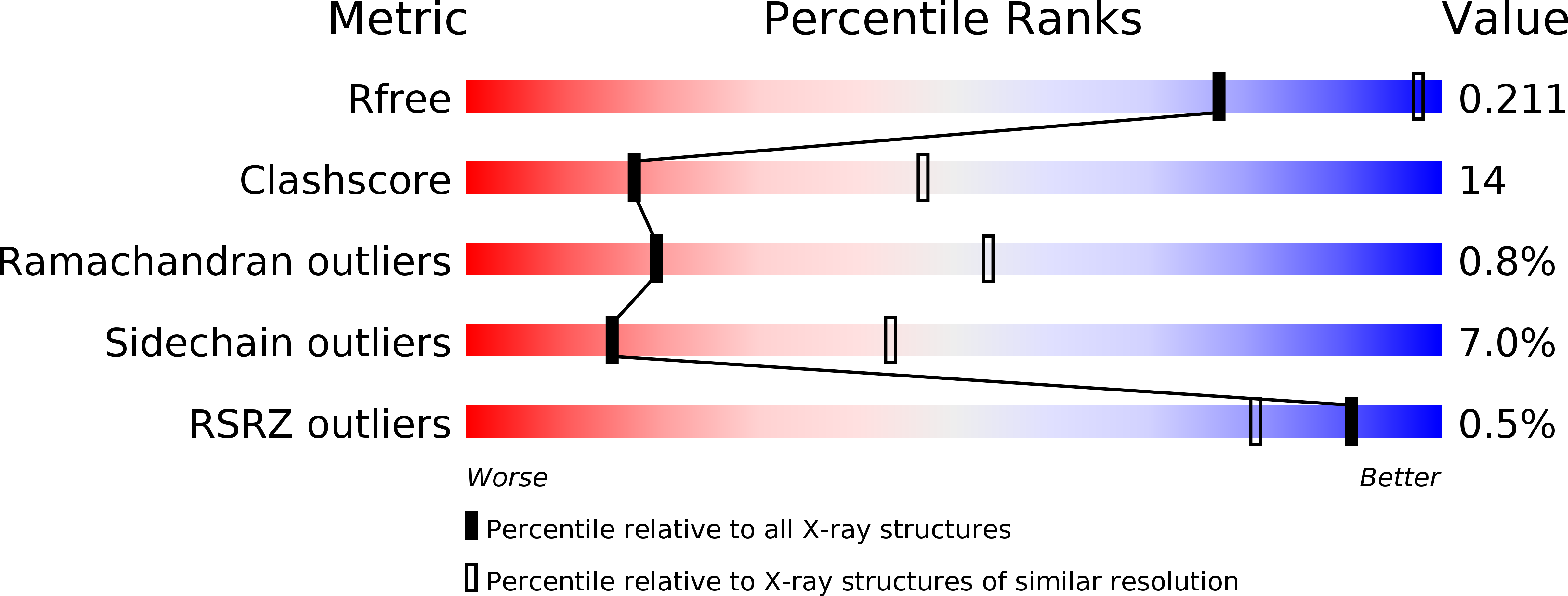
Resolution:
2.95 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.18
Space Group:
P 65 2 2