Deposition Date
2010-11-24
Release Date
2011-06-15
Last Version Date
2023-12-20
Entry Detail
PDB ID:
2XZ9
Keywords:
Title:
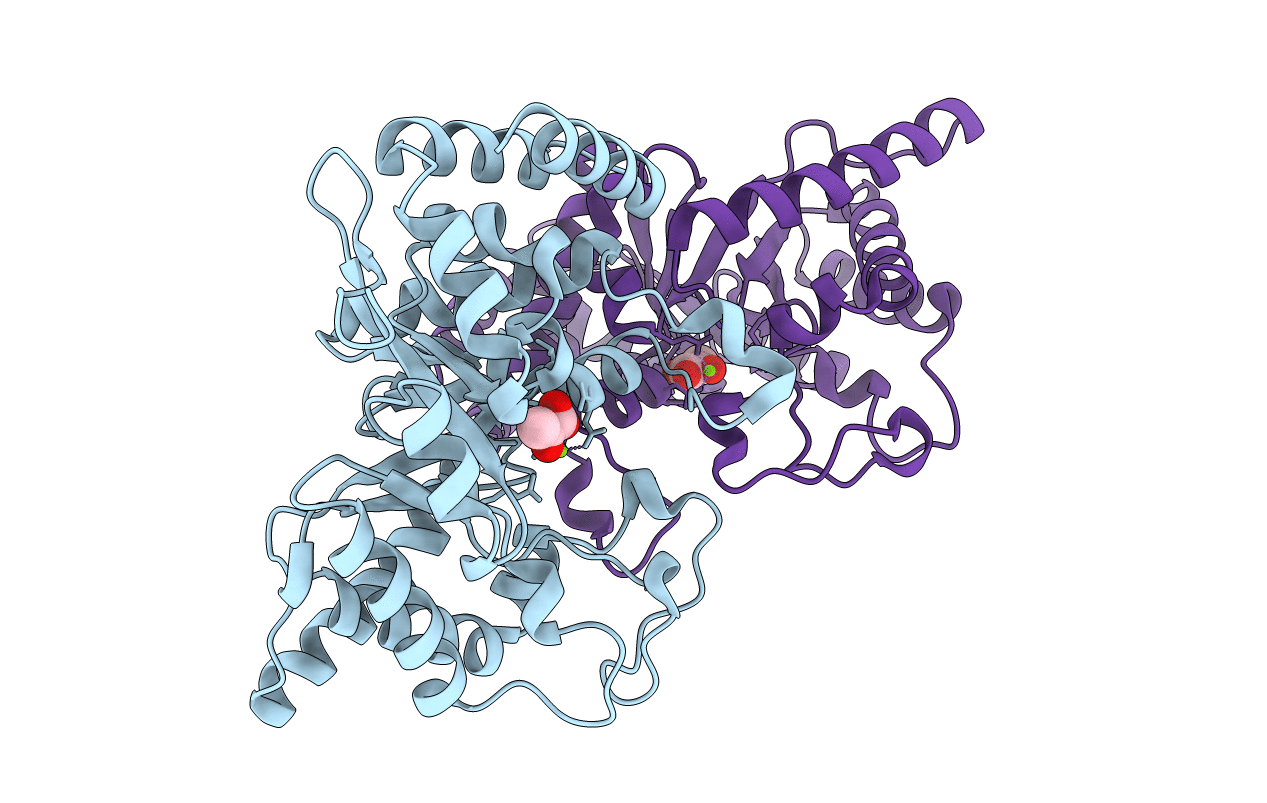
CRYSTAL STRUCTURE FROM THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS)
Biological Source:
Source Organism(s):
THERMOANAEROBACTER TENGCONGENSIS (Taxon ID: 119072)
Expression System(s):
Method Details:
Experimental Method:
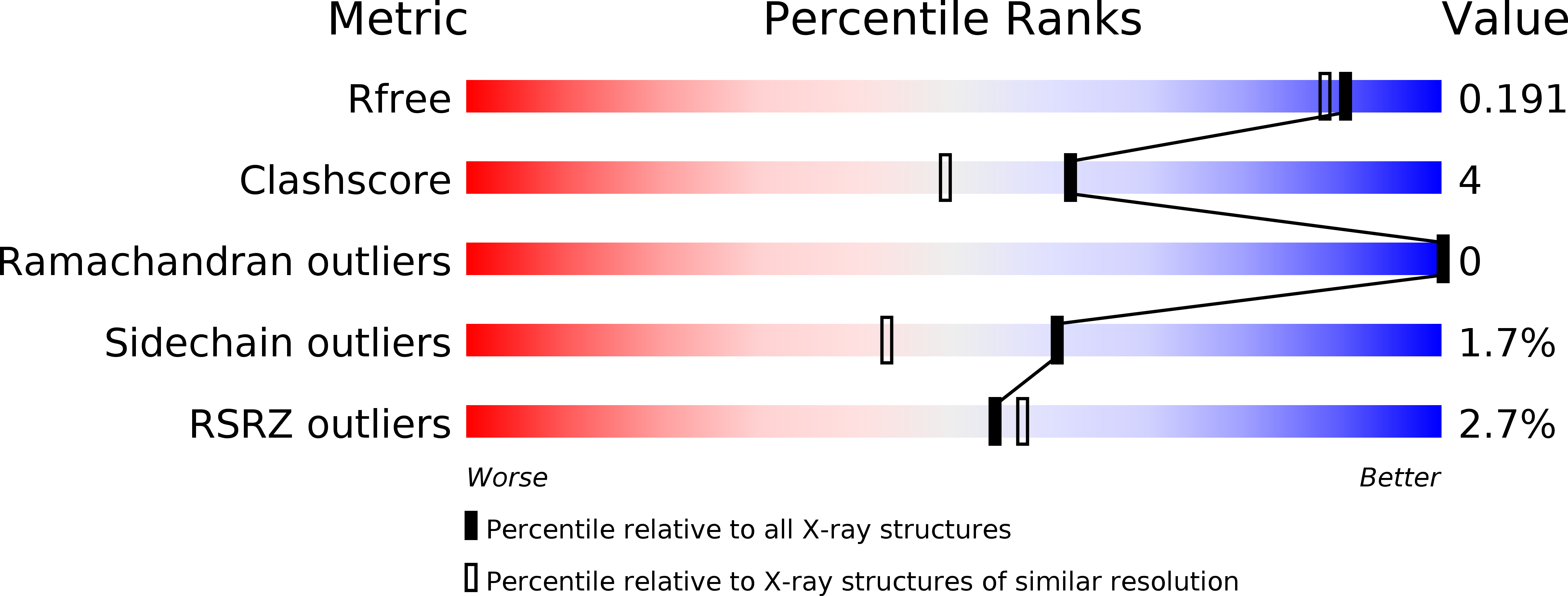
Resolution:
1.68 Å
R-Value Free:
0.19
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21