Deposition Date
2010-11-16
Release Date
2011-06-29
Last Version Date
2024-05-15
Entry Detail
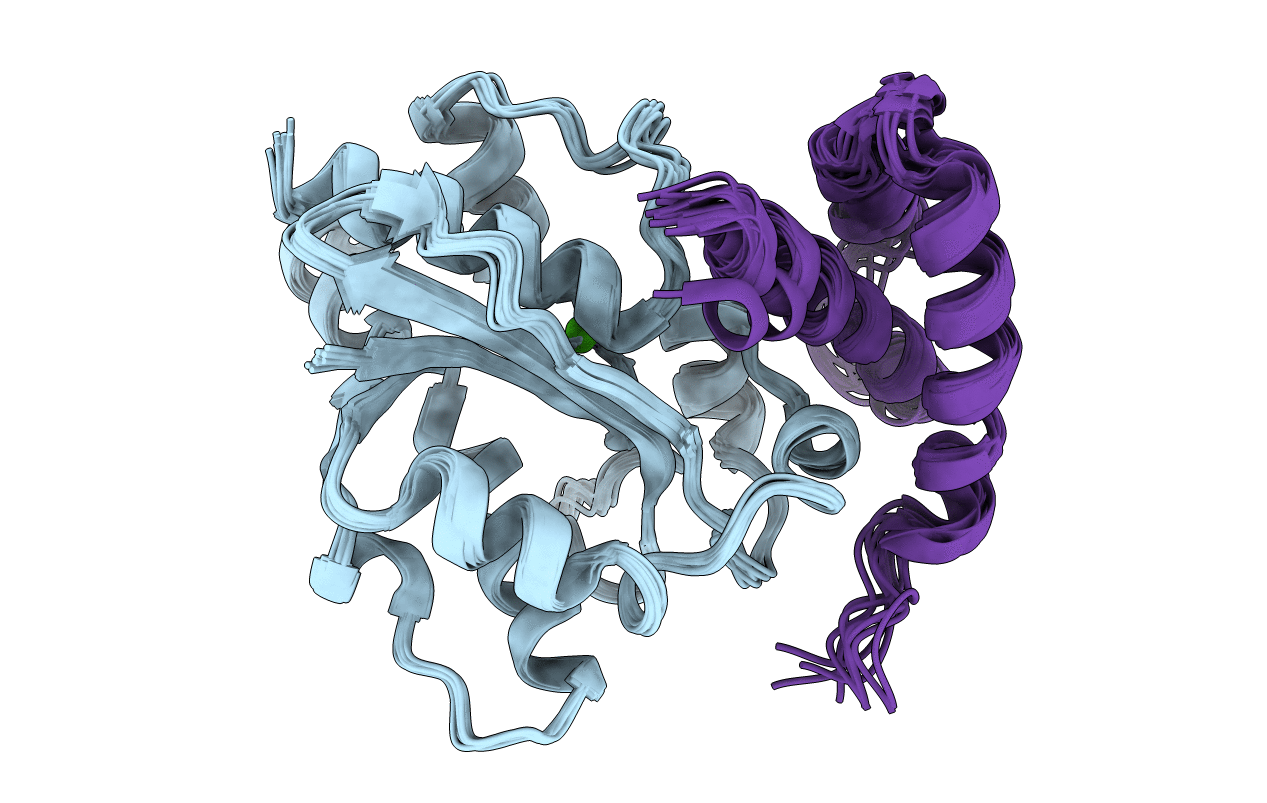
PDB ID:
2XY8
Keywords:
Title:
Paramagnetic-based NMR structure of the complex between the N- terminal epsilon domain and the theta domain of the DNA polymerase III
Biological Source:
Source Organism(s):
ESCHERICHIA COLI (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
10
Selection Criteria:
TOP 10 STRUCTURES OF THE BEST CLUSTER


