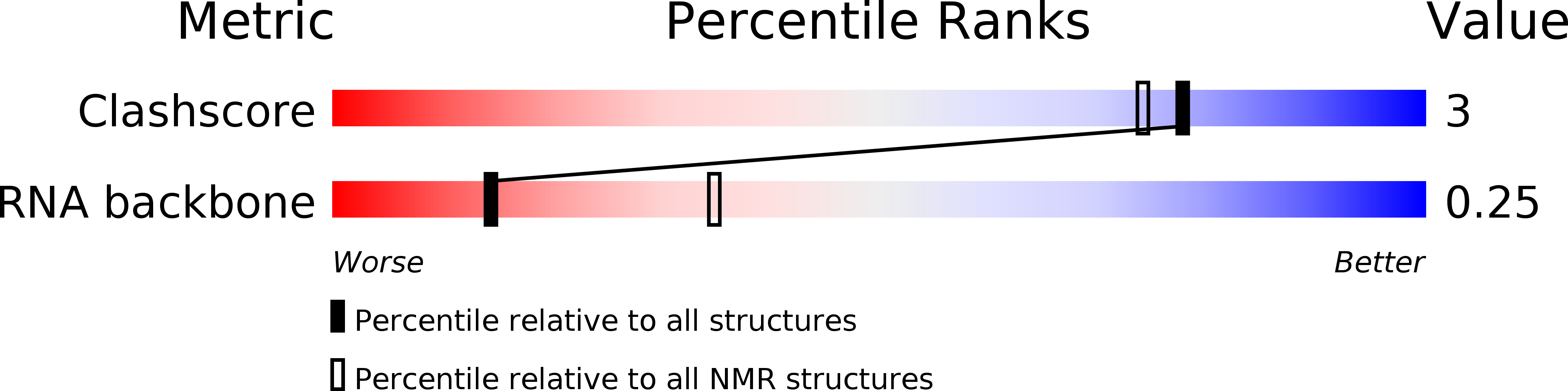
Deposition Date
2010-05-12
Release Date
2010-05-26
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2XEB
Keywords:
Title:
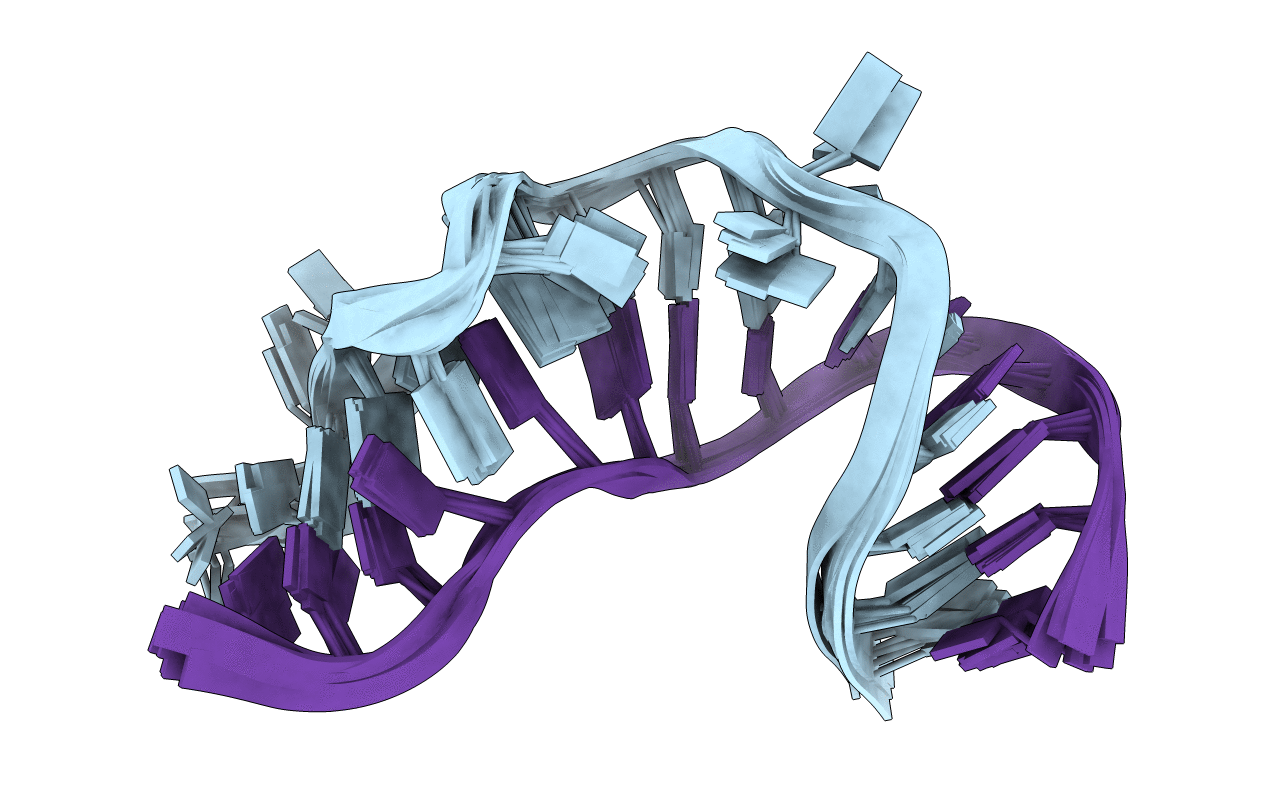
NMR STRUCTURE OF THE PROTEIN-UNBOUND SPLICEOSOMAL U4 SNRNA 5' STEM LOOP
Biological Source:
Source Organism(s):
HOMO SAPIENS (Taxon ID: 9606)
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
10
Selection Criteria:
LOWEST ENERGY STRUCTURES