Deposition Date
2010-04-30
Release Date
2010-09-22
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2XDF
Keywords:
Title:
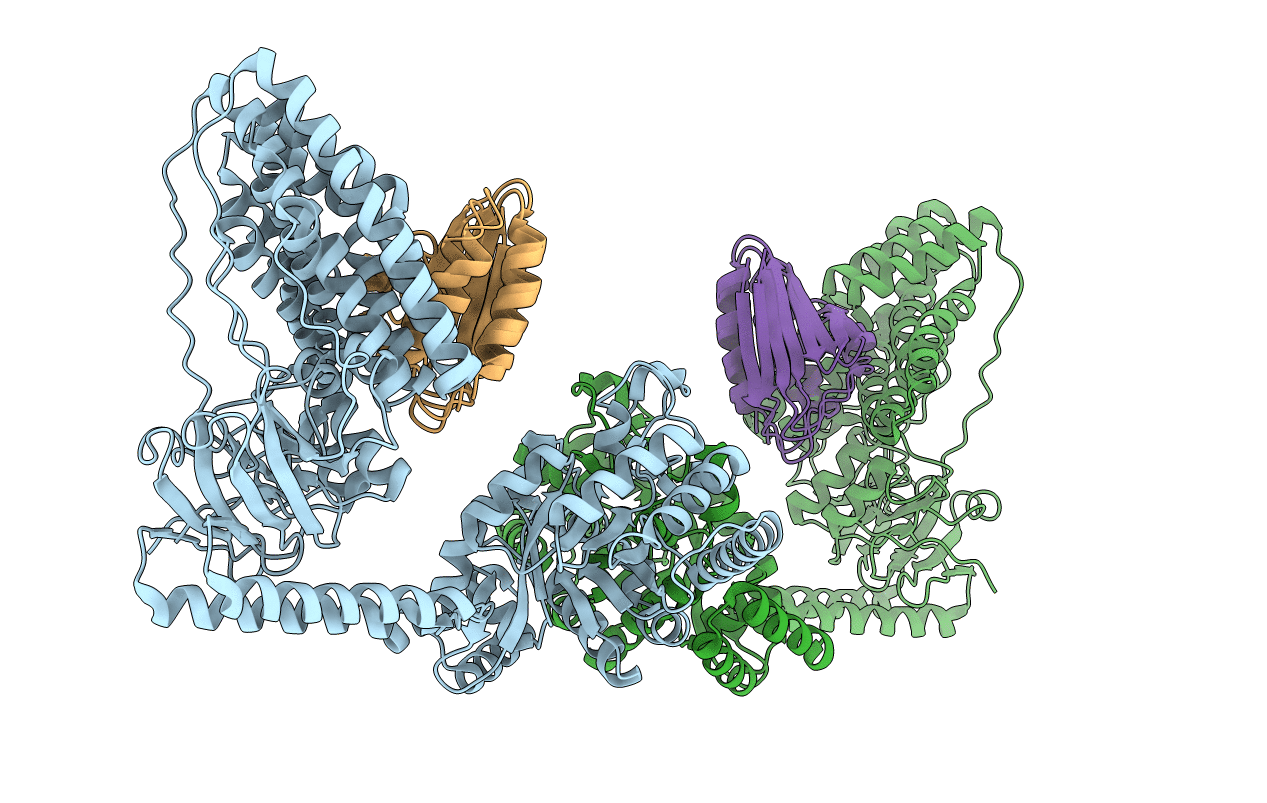
Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering
Biological Source:
Source Organism(s):
ESCHERICHIA COLI (Taxon ID: 469008)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
120
Conformers Submitted:
2
Selection Criteria:
BEST EXPERIMENT FIT, AND THEN LOWEST ENERGY


