Deposition Date
2010-04-24
Release Date
2010-08-04
Last Version Date
2023-12-20
Entry Detail
PDB ID:
2XCQ
Keywords:
Title:
The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase
Biological Source:
Source Organism(s):
STAPHYLOCOCCUS AUREUS (Taxon ID: 158879)
Expression System(s):
Method Details:
Experimental Method:
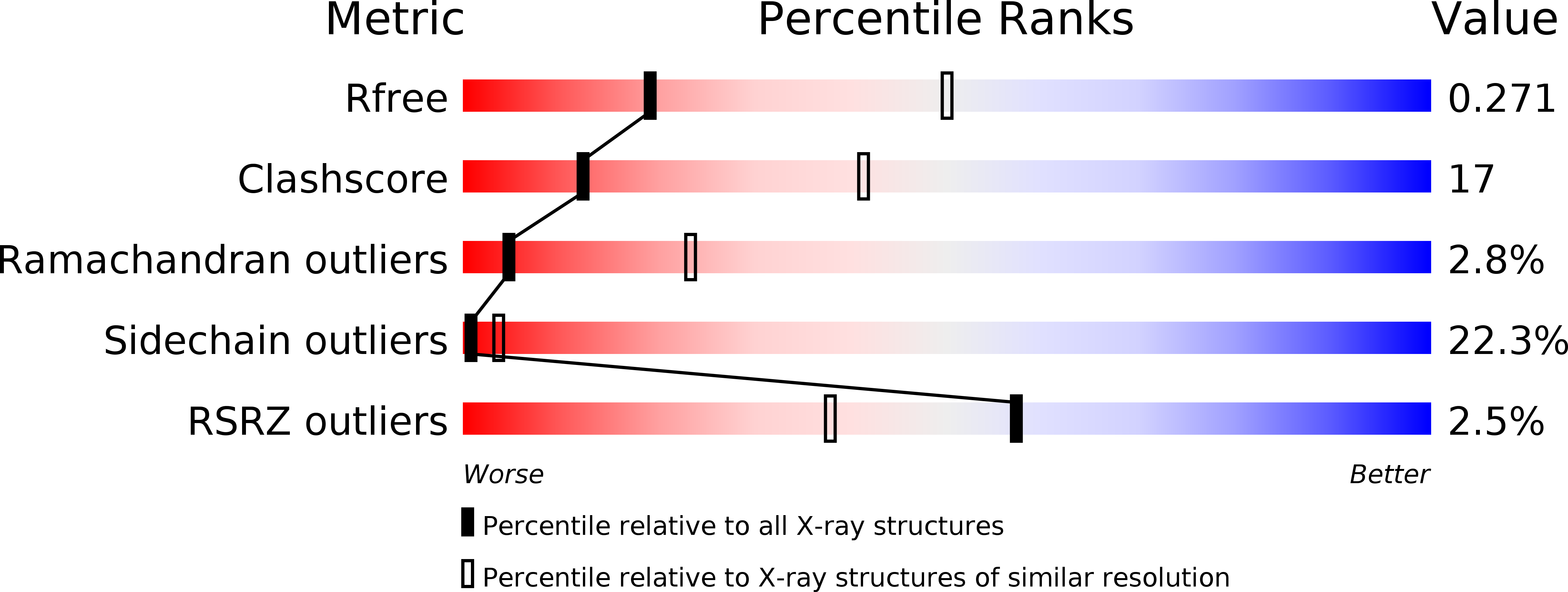
Resolution:
2.98 Å
R-Value Free:
0.28
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 61 2 2