Deposition Date
2010-02-17
Release Date
2010-04-07
Last Version Date
2023-12-20
Entry Detail
PDB ID:
2X6J
Keywords:
Title:
THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-93
Biological Source:
Source Organism(s):
DROSOPHILA MELANOGASTER (Taxon ID: 7227)
Expression System(s):
Method Details:
Experimental Method:
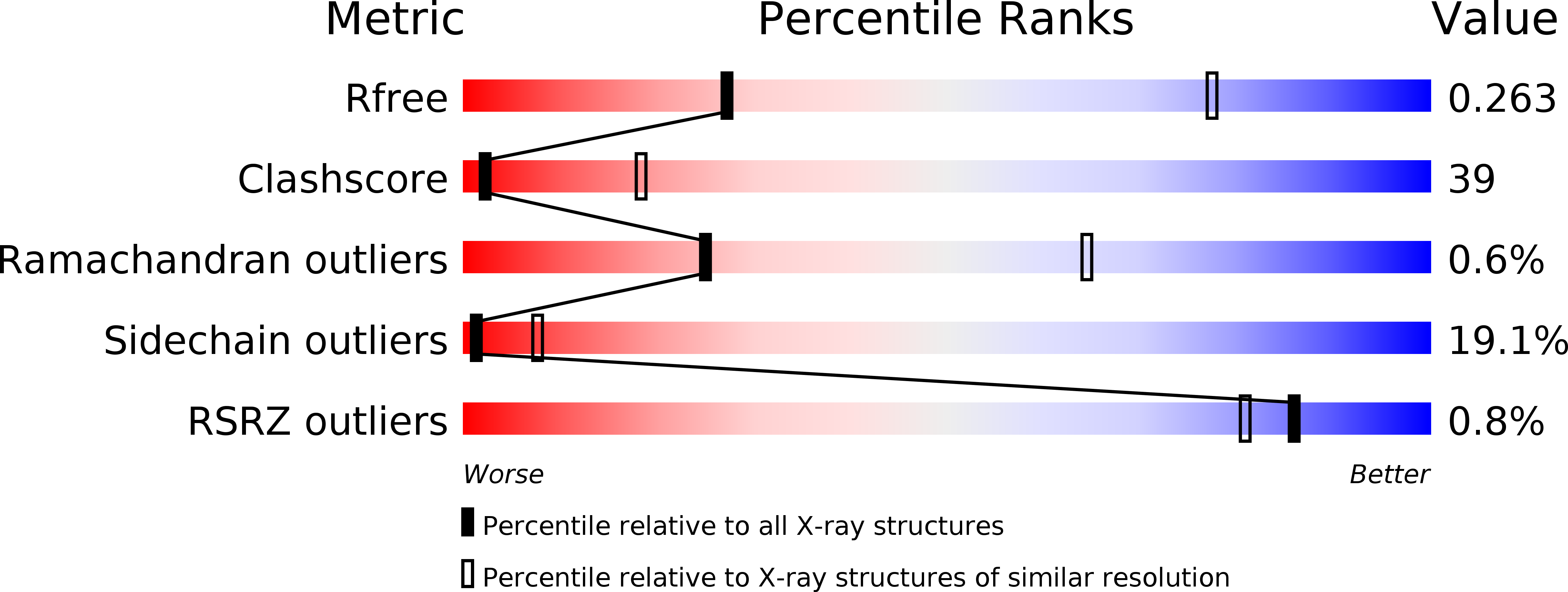
Resolution:
3.50 Å
R-Value Free:
0.27
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
I 21 21 21