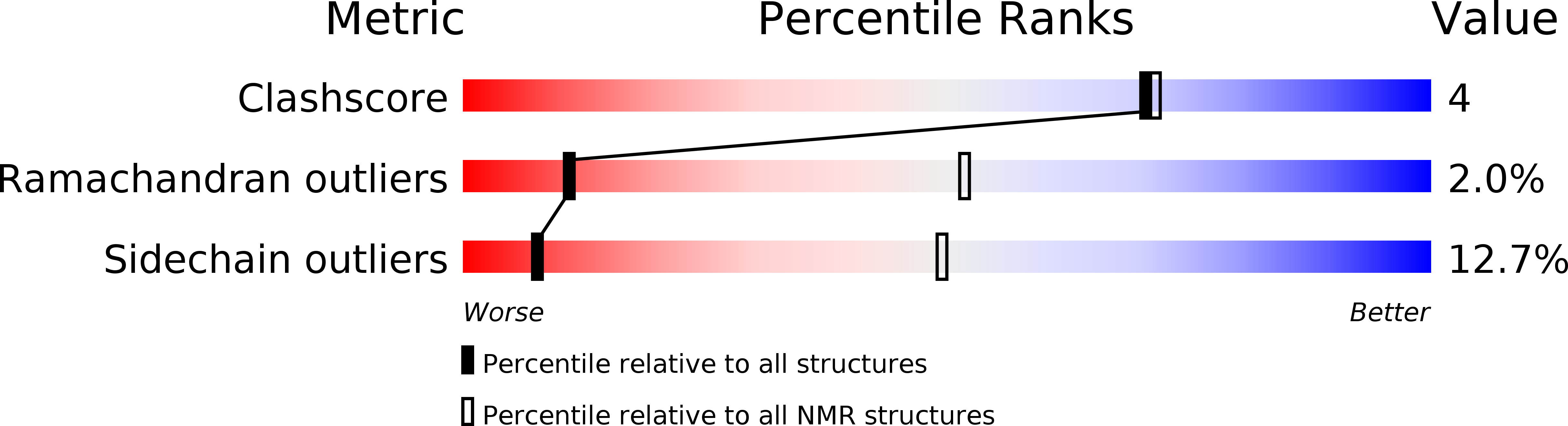Deposition Date
2007-10-26
Release Date
2008-01-08
Last Version Date
2024-11-13
Entry Detail
PDB ID:
2VER
Keywords:
Title:
Structural model for the complex between the Dr adhesins and carcinoembryonic antigen (CEA)
Biological Source:
Source Organism(s):
ESCHERICHIA COLI (Taxon ID: 562)
HOMO SAPIENS (Taxon ID: 9606)
HOMO SAPIENS (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
1
Selection Criteria:
REPRESENTATIVE STRUCTURE