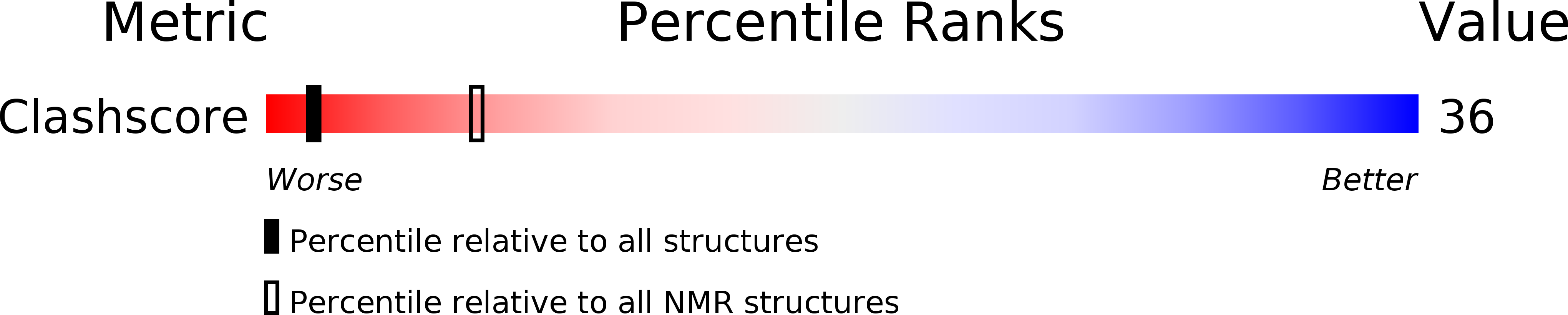Deposition Date
2007-06-18
Release Date
2007-10-09
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2V3L
Keywords:
Title:
Orientational and dynamical heterogeneity of Rhodamine 6G terminally attached to a DNA helix
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Conformers Submitted:
2