Deposition Date
2007-03-28
Release Date
2007-07-03
Last Version Date
2023-12-13
Entry Detail
PDB ID:
2UXK
Keywords:
Title:
X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 10 in the charge-separated state
Biological Source:
Source Organism(s):
RHODOBACTER SPHAEROIDES (Taxon ID: 1063)
Method Details:
Experimental Method:
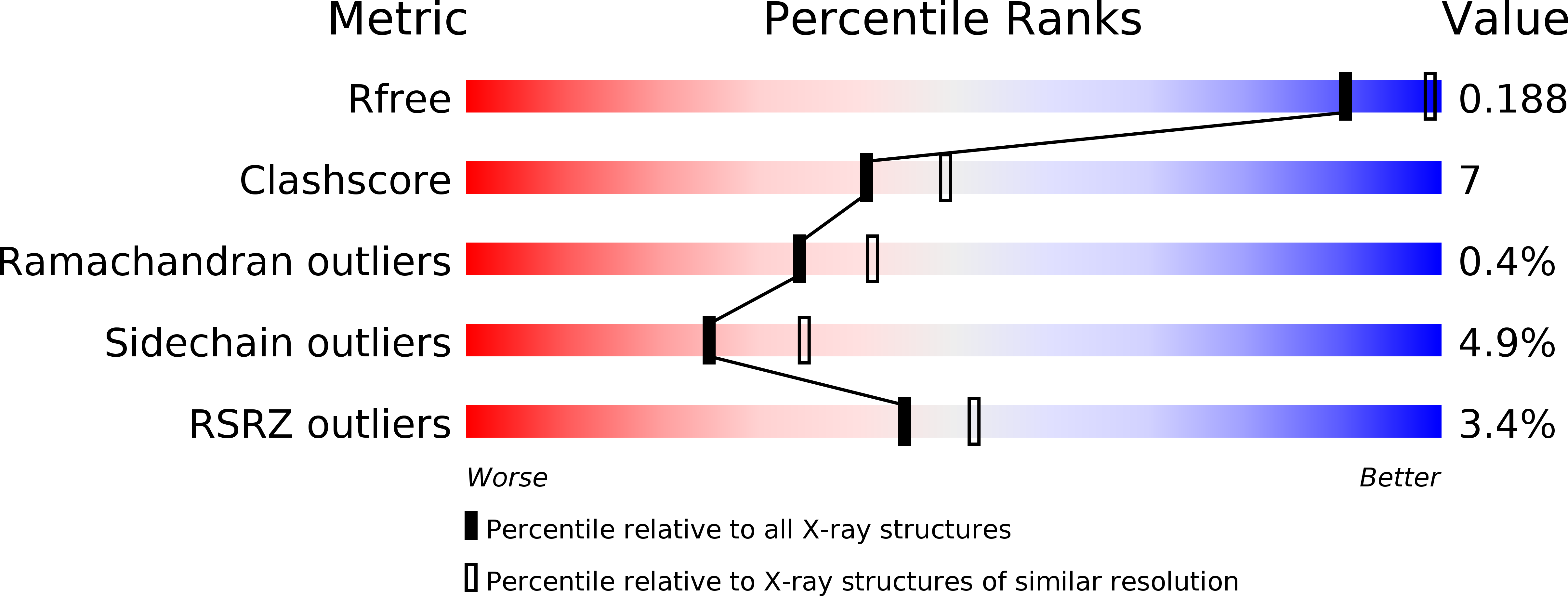
Resolution:
2.31 Å
R-Value Free:
0.21
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 43 21 2