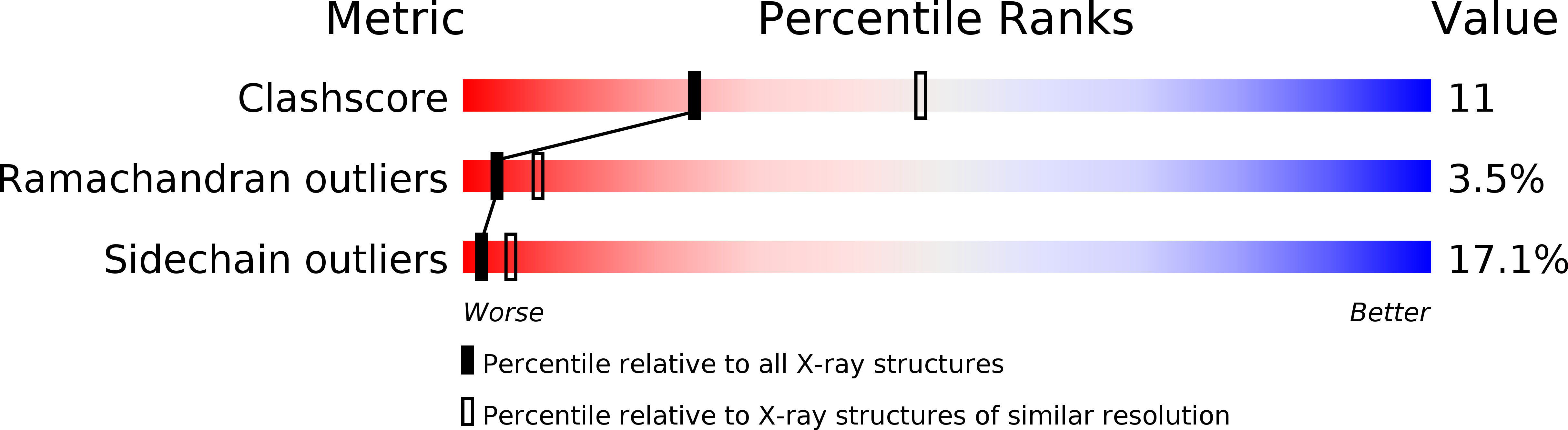Deposition Date
1993-04-05
Release Date
1993-07-15
Last Version Date
2024-02-21
Entry Detail
PDB ID:
2TDD
Keywords:
Title:
STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE
Biological Source:
Source Organism(s):
Lactobacillus casei (Taxon ID: 1582)
Method Details: