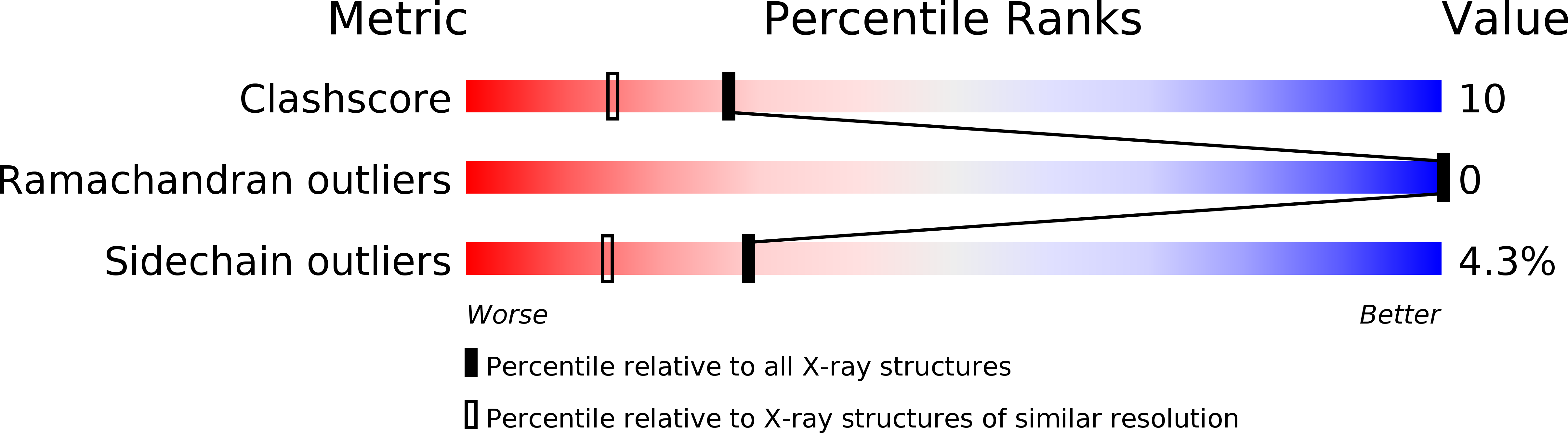Deposition Date
1994-01-14
Release Date
1994-04-30
Last Version Date
2024-12-25
Entry Detail
PDB ID:
2TBS
Keywords:
Title:
COLD-ADAPTION OF ENZYMES: STRUCTURAL COMPARISON BETWEEN SALMON AND BOVINE TRYPSINS
Biological Source:
Source Organism(s):
Salmo salar (Taxon ID: 8030)
Method Details: