Deposition Date
1998-12-10
Release Date
2000-01-19
Last Version Date
2024-11-13
Entry Detail
PDB ID:
2STA
Keywords:
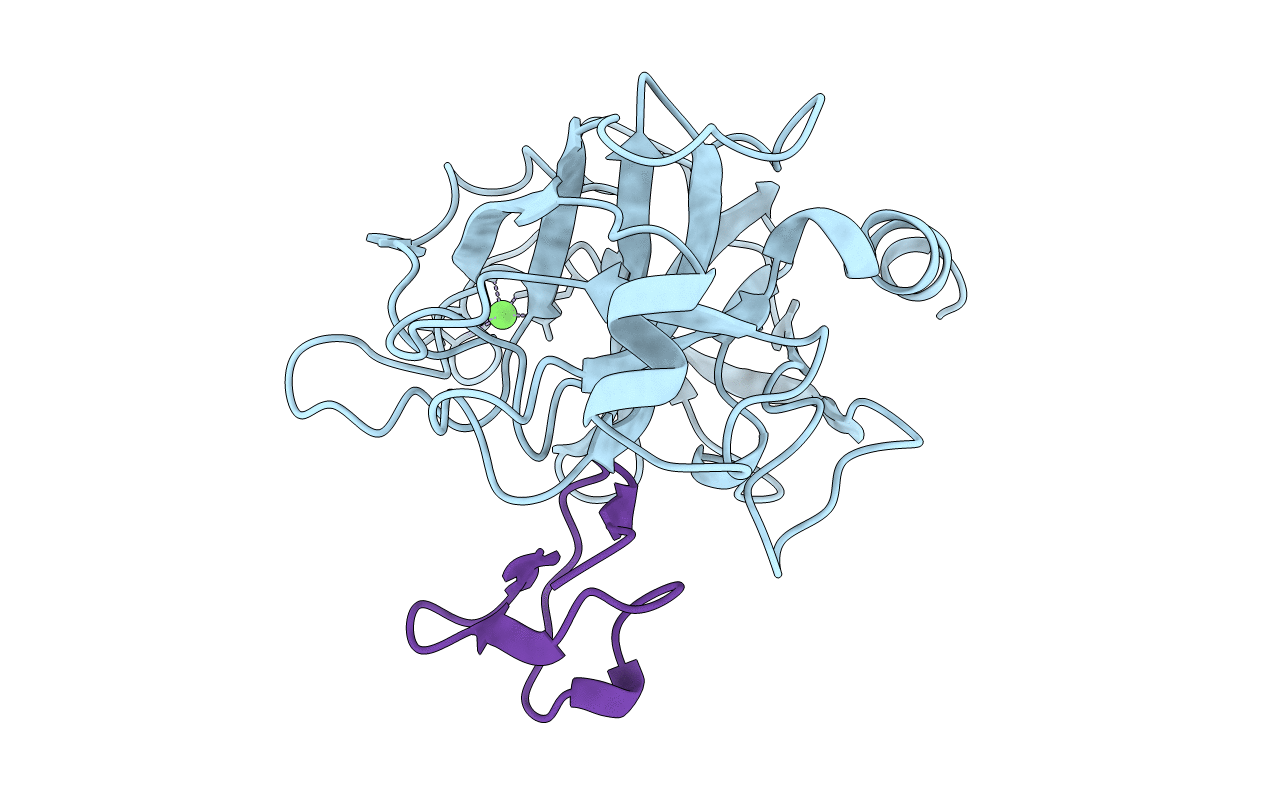
Title:
ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I)
Biological Source:
Source Organism(s):
Salmo salar (Taxon ID: 8030)
Cucurbita maxima (Taxon ID: 3661)
Cucurbita maxima (Taxon ID: 3661)
Method Details:
Experimental Method:
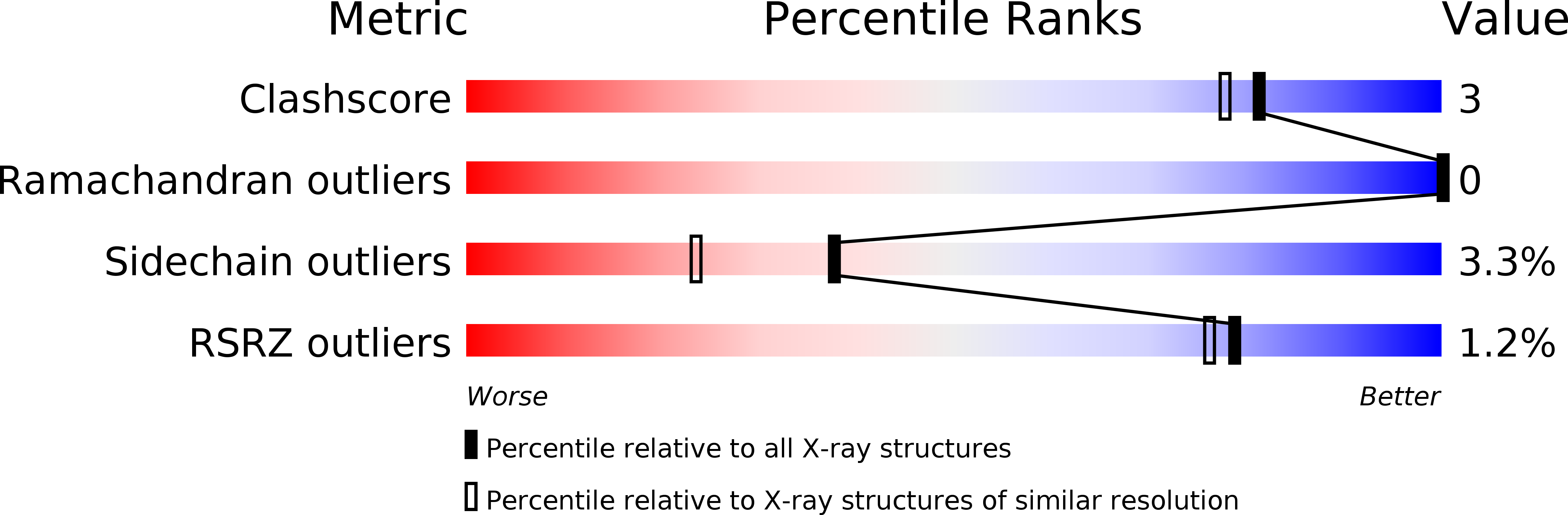
Resolution:
1.80 Å
R-Value Free:
0.23
R-Value Work:
0.19
Space Group:
P 21 21 21