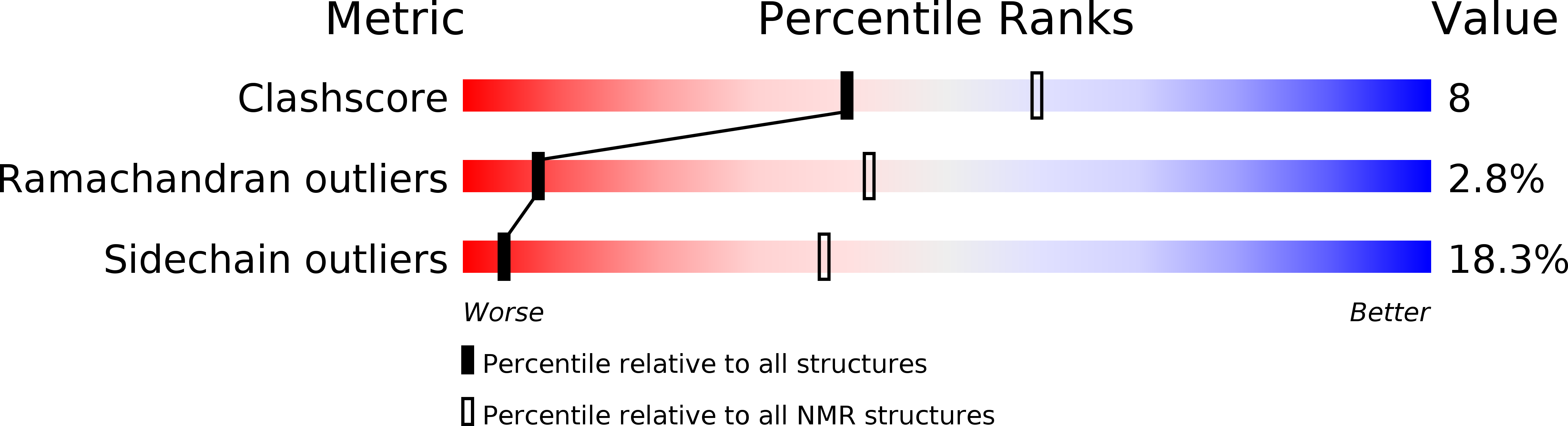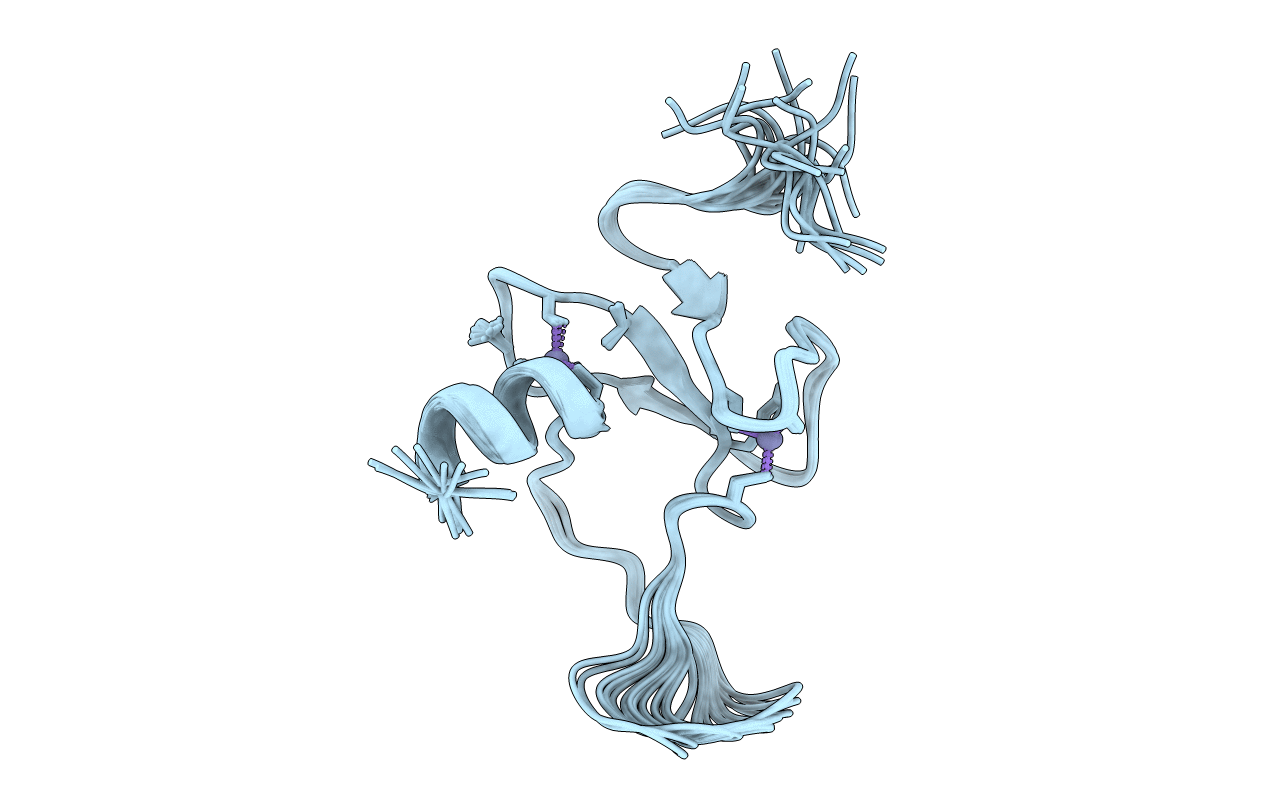
Deposition Date
2012-01-12
Release Date
2012-08-15
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2RSD
Keywords:
Title:
Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice
Biological Source:
Source Organism(s):
Oryza sativa Japonica Group (Taxon ID: 39947)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the least restraint violations