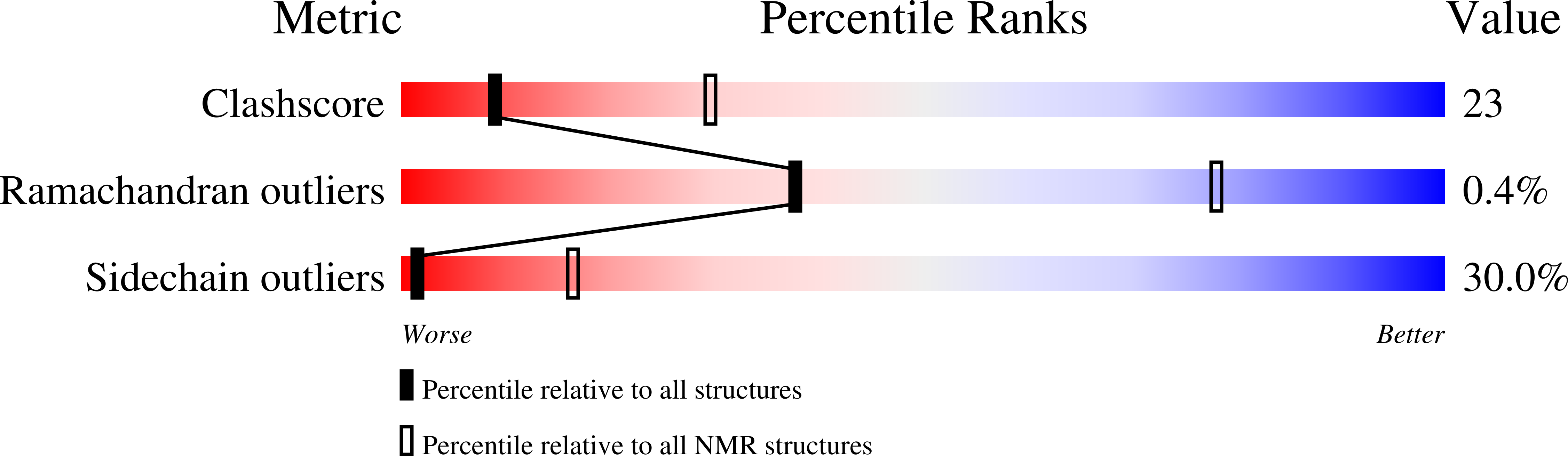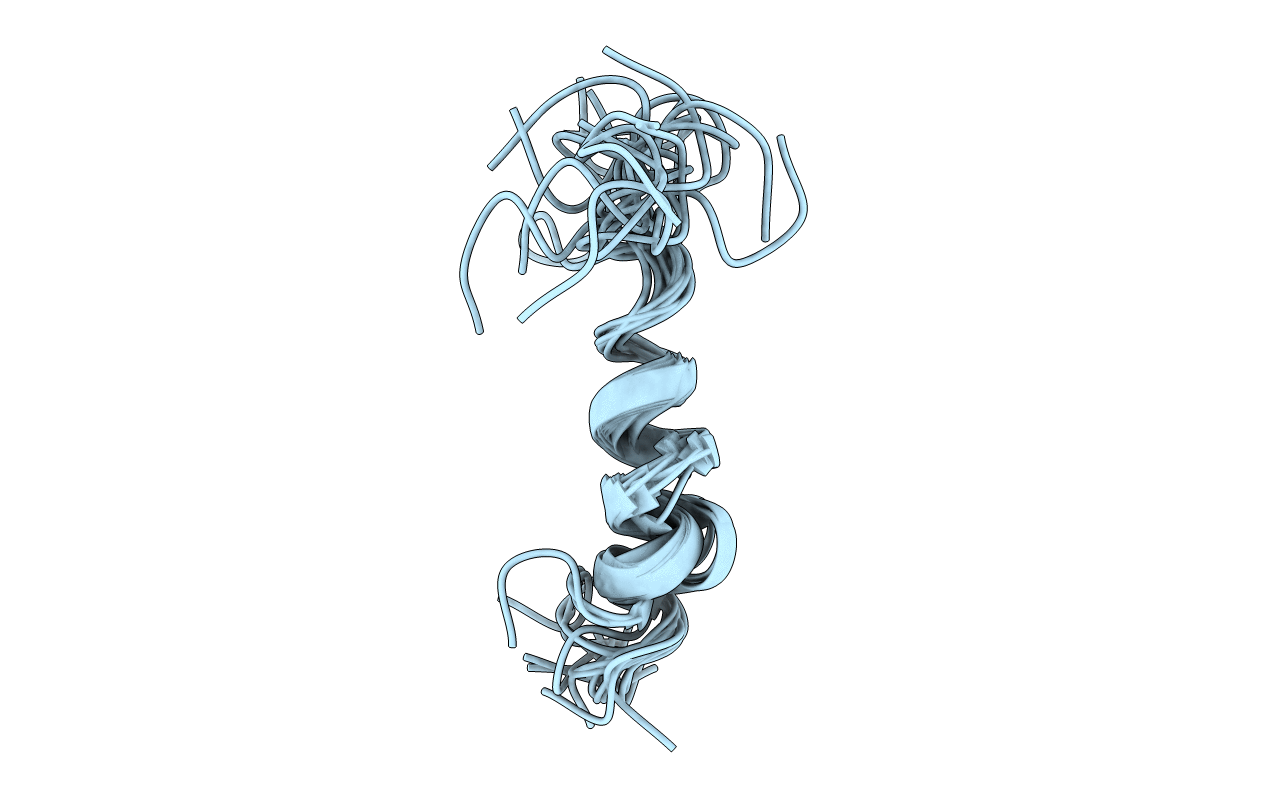
Deposition Date
2008-11-08
Release Date
2009-03-24
Last Version Date
2024-05-29
Entry Detail
PDB ID:
2RPW
Keywords:
Title:
Structure of a peptide derived from H+-V-ATPase subunit a
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy