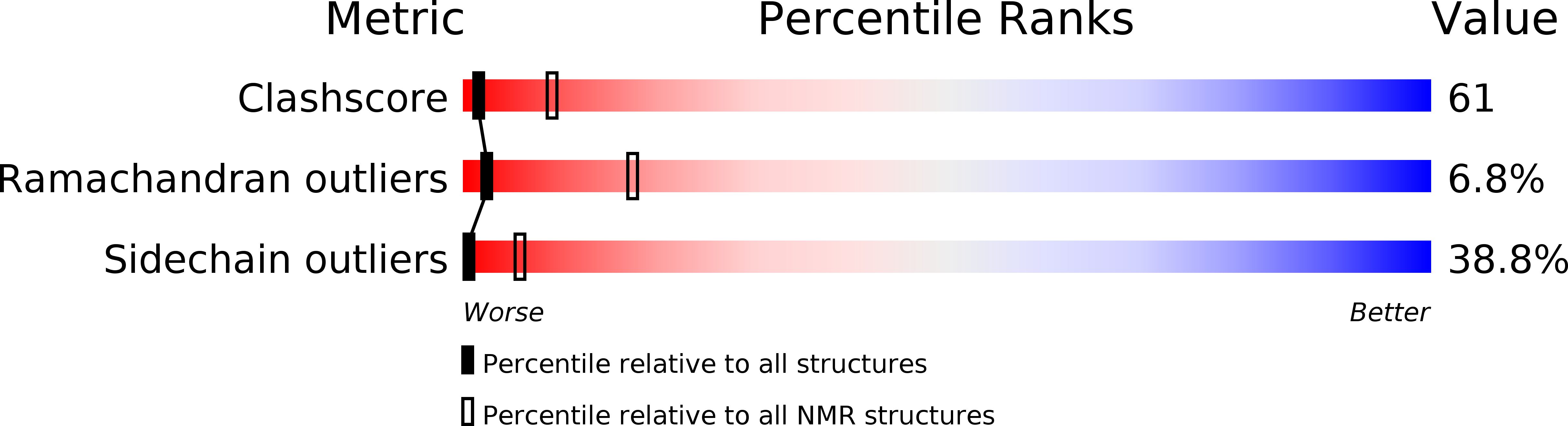
Deposition Date
1994-08-19
Release Date
1995-01-26
Last Version Date
2024-11-20
Entry Detail
PDB ID:
2PLE
Keywords:
Title:
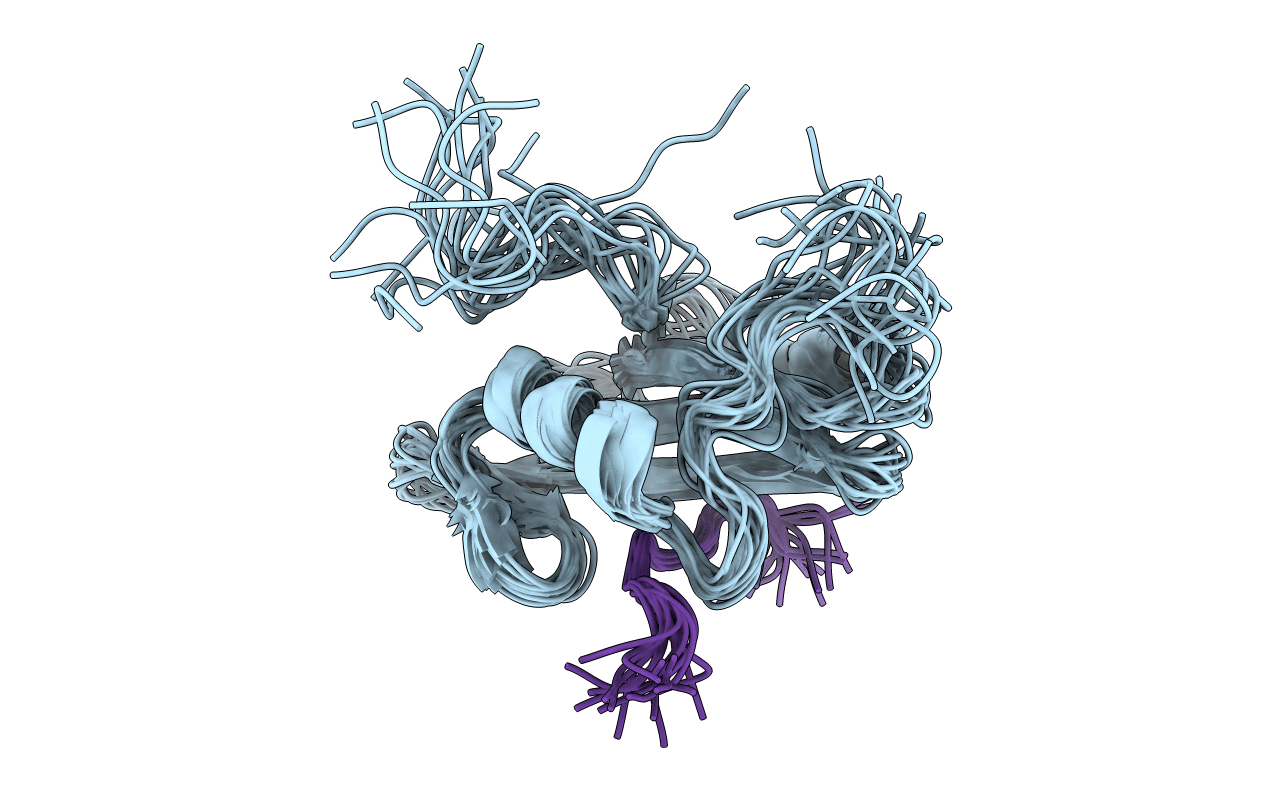
NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE
Biological Source:
Source Organism(s):
Bos taurus (Taxon ID: 9913)
Method Details:
Experimental Method:
Conformers Submitted:
18