Deposition Date
2007-03-20
Release Date
2007-05-29
Last Version Date
2024-10-30
Entry Detail
PDB ID:
2P7S
Keywords:
Title:
Enzymatic and Structural Characterisation of Amphinase, a Novel Cytotoxic Ribonuclease from Rana pipiens Oocytes
Biological Source:
Source Organism(s):
Rana pipiens (Taxon ID: 8404)
Method Details:
Experimental Method:
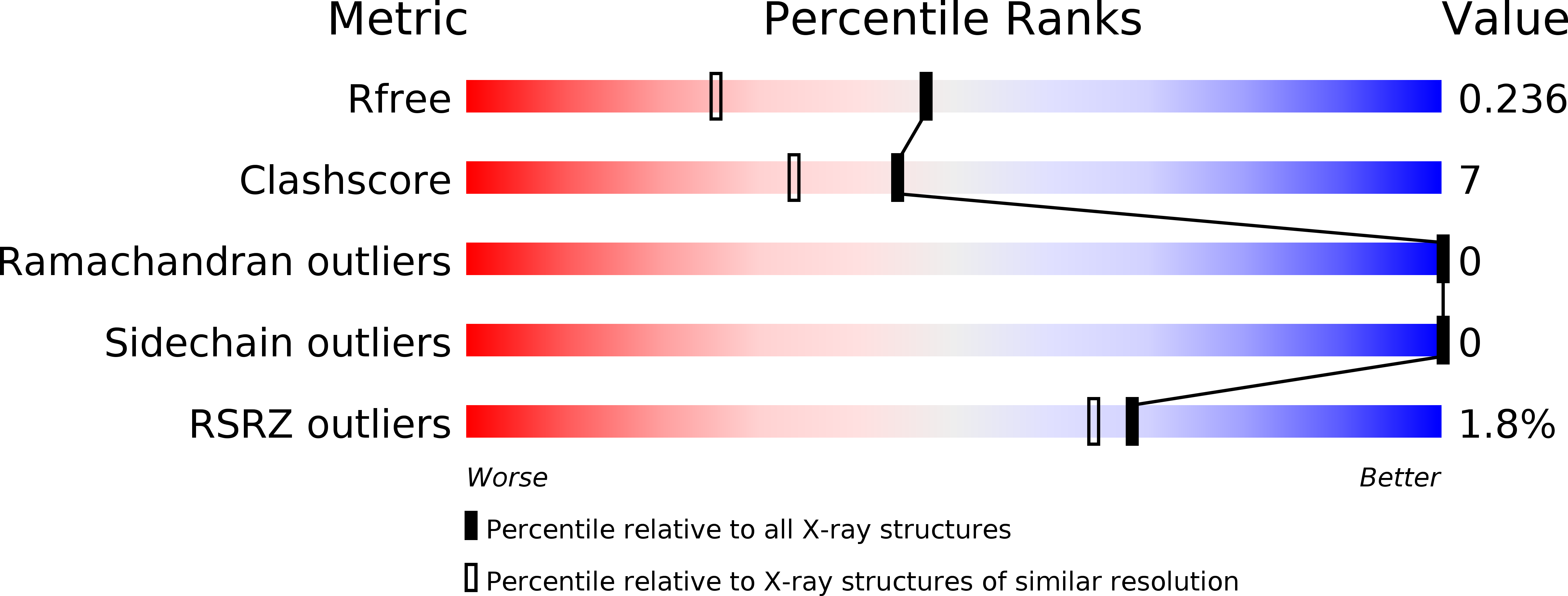
Resolution:
1.80 Å
R-Value Free:
0.24
R-Value Work:
0.22
Space Group:
P 41