Deposition Date
2007-03-11
Release Date
2008-03-11
Last Version Date
2024-11-06
Entry Detail
PDB ID:
2P45
Keywords:
Title:
Complex of a camelid single-domain vhh antibody fragment with RNASE A at 1.1A resolution: SE5B-ORTHO-1 crystal form with five se-met sites (L4M, M34, M51, F68M, M83) in vhh scaffold.
Biological Source:
Source Organism(s):
Camelus dromedarius (Taxon ID: 9838)
Bos taurus (Taxon ID: 9913)
Bos taurus (Taxon ID: 9913)
Expression System(s):
Method Details:
Experimental Method:
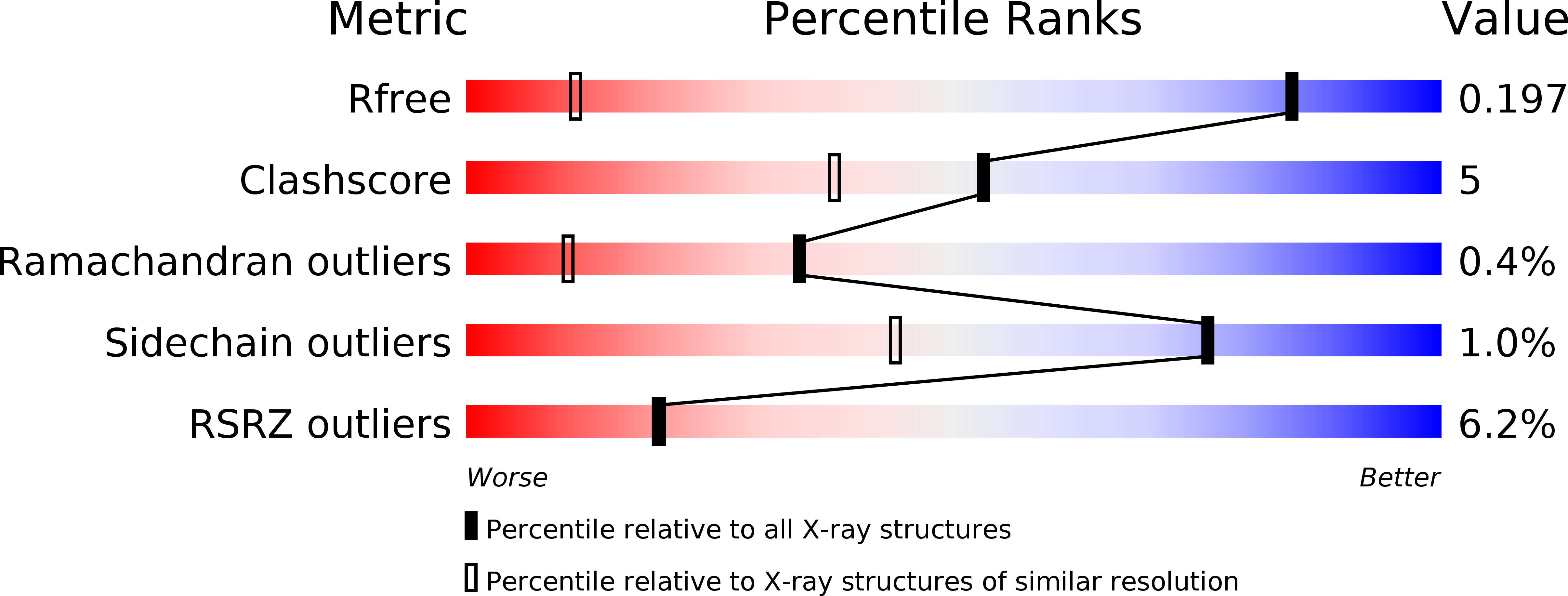
Resolution:
1.10 Å
R-Value Free:
0.19
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21