Deposition Date
2007-01-21
Release Date
2008-01-15
Last Version Date
2023-12-27
Entry Detail
PDB ID:
2OM7
Keywords:
Title:
Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors
Biological Source:
Source Organism(s):
Thermus thermophilus (Taxon ID: 274)
Expression System(s):
Method Details:
Experimental Method:
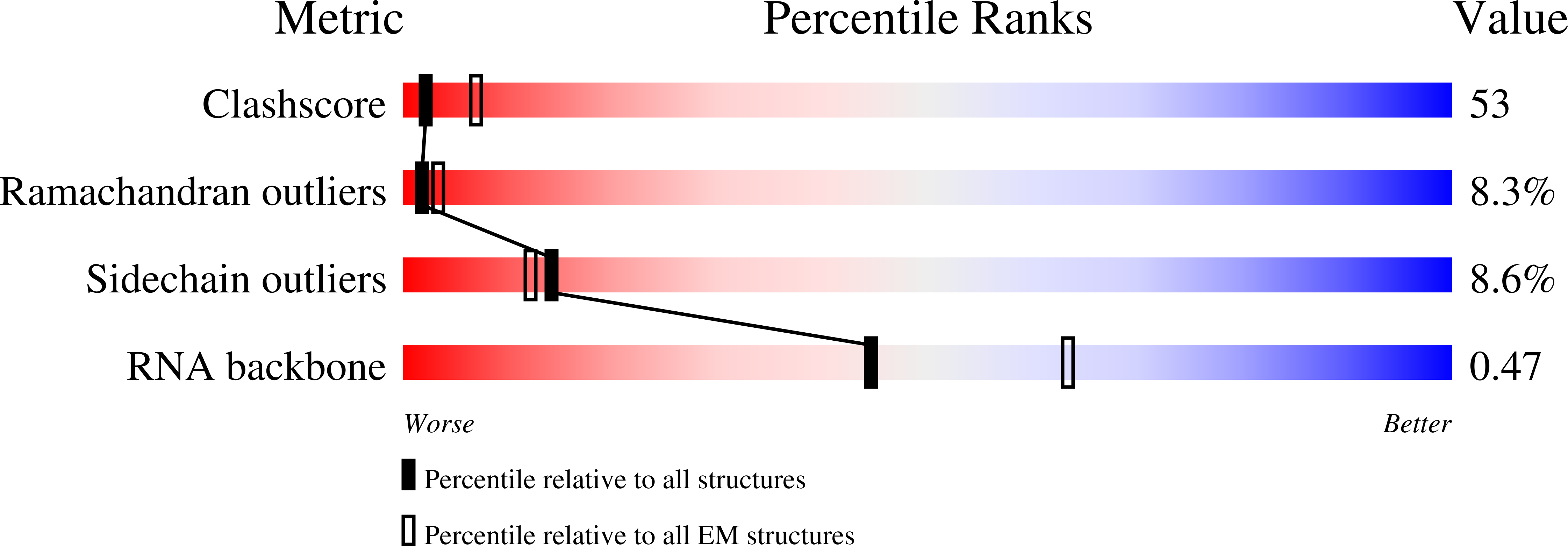
Resolution:
7.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE