Deposition Date
2006-12-02
Release Date
2007-11-06
Last Version Date
2023-12-27
Entry Detail
PDB ID:
2O3V
Keywords:
Title:
Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33
Method Details:
Experimental Method:
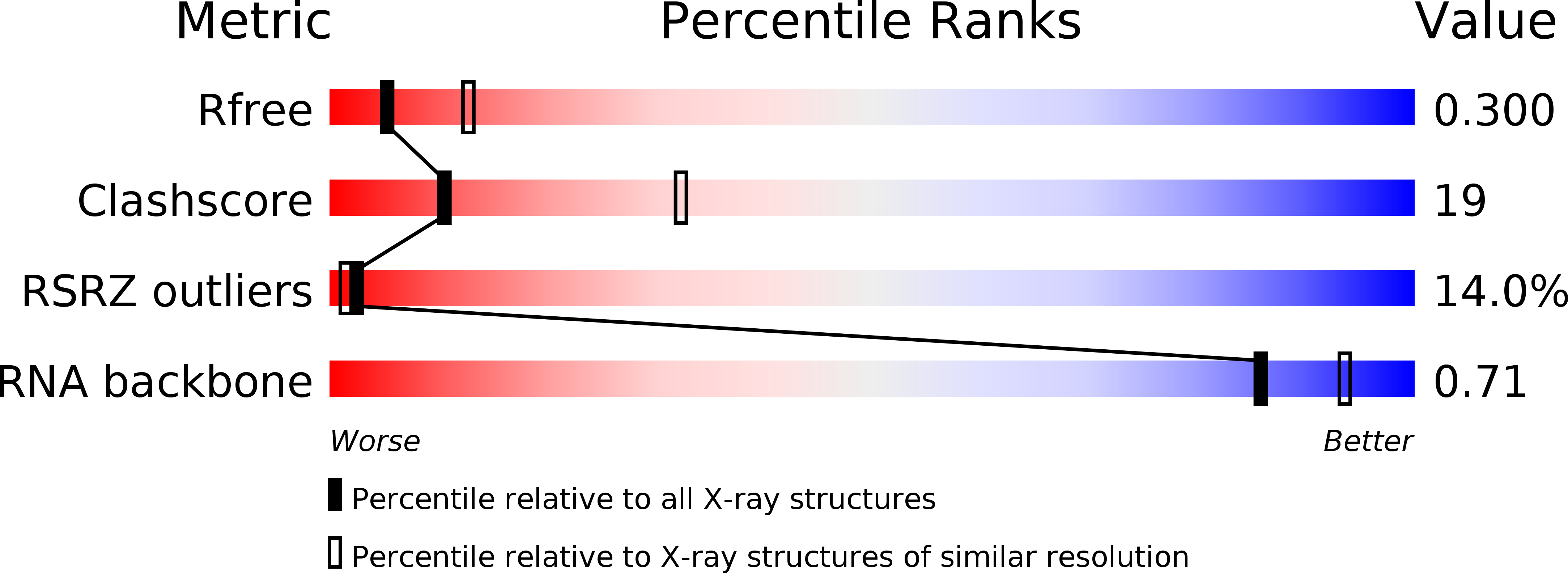
Resolution:
2.80 Å
R-Value Free:
0.29
R-Value Work:
0.23
Space Group:
P 1 21 1