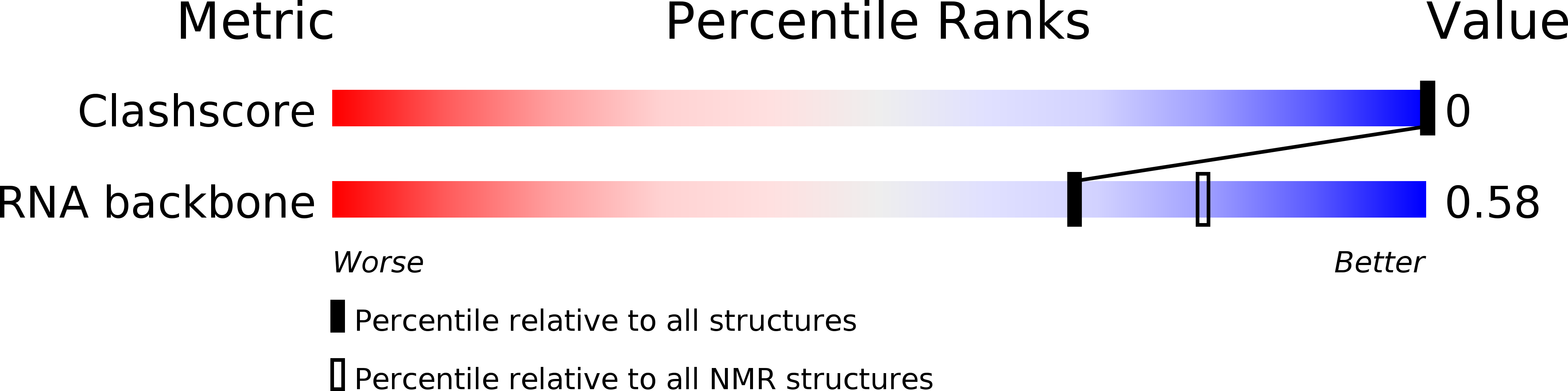
Deposition Date
2015-06-23
Release Date
2015-12-02
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2N4L
Keywords:
Title:
Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1
Method Details:
Experimental Method:
Conformers Calculated:
256
Conformers Submitted:
10
Selection Criteria:
structures with the least restraint violations