Deposition Date
2015-06-17
Release Date
2015-07-01
Last Version Date
2024-05-15
Entry Detail
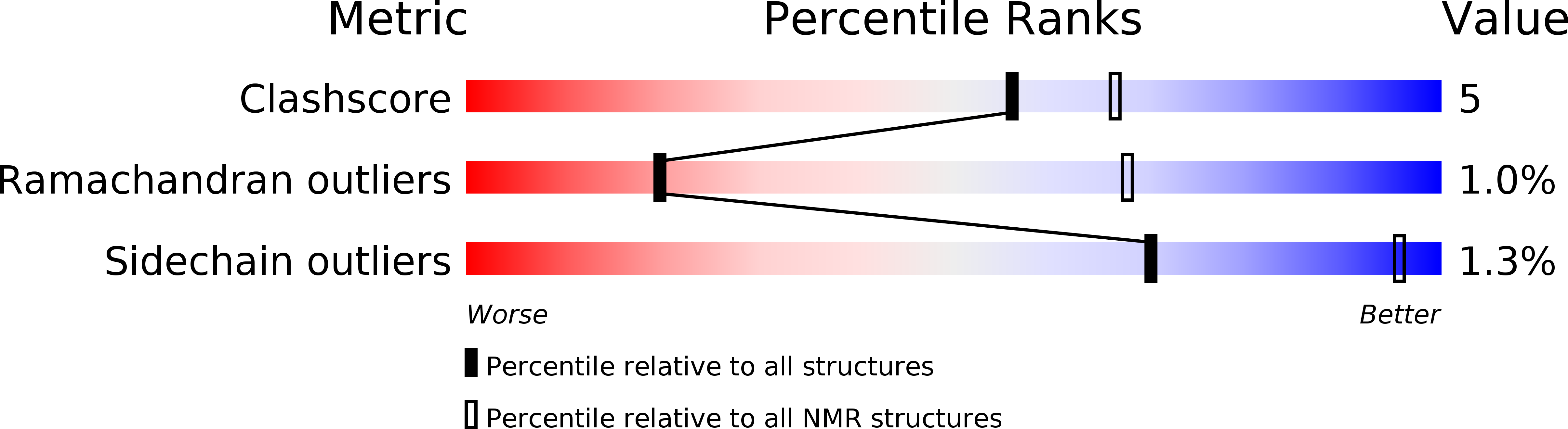
PDB ID:
2N45
Keywords:
Title:
EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy