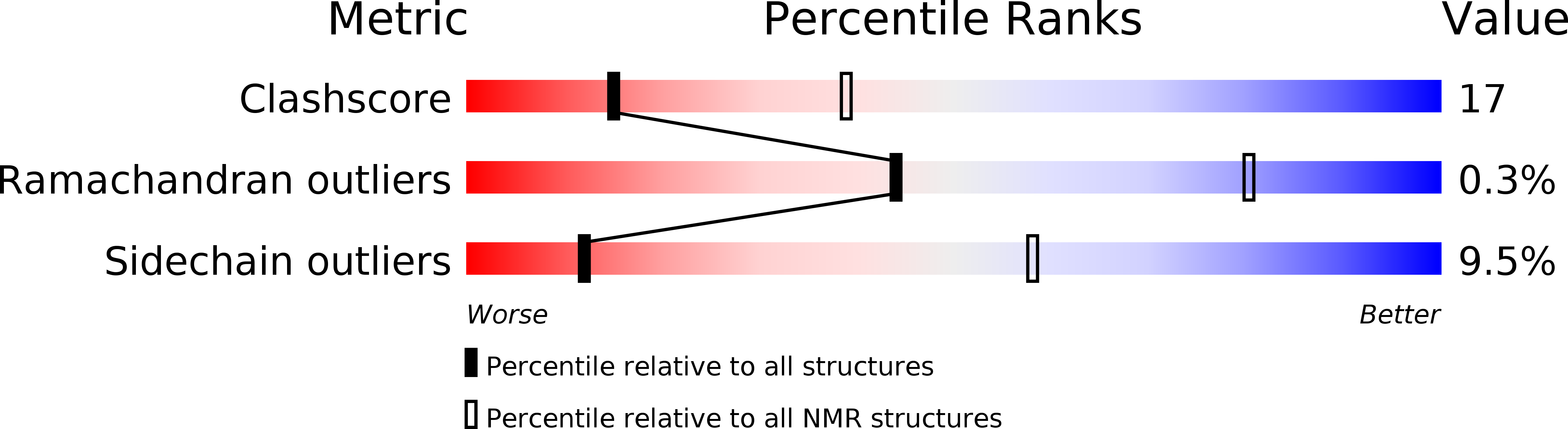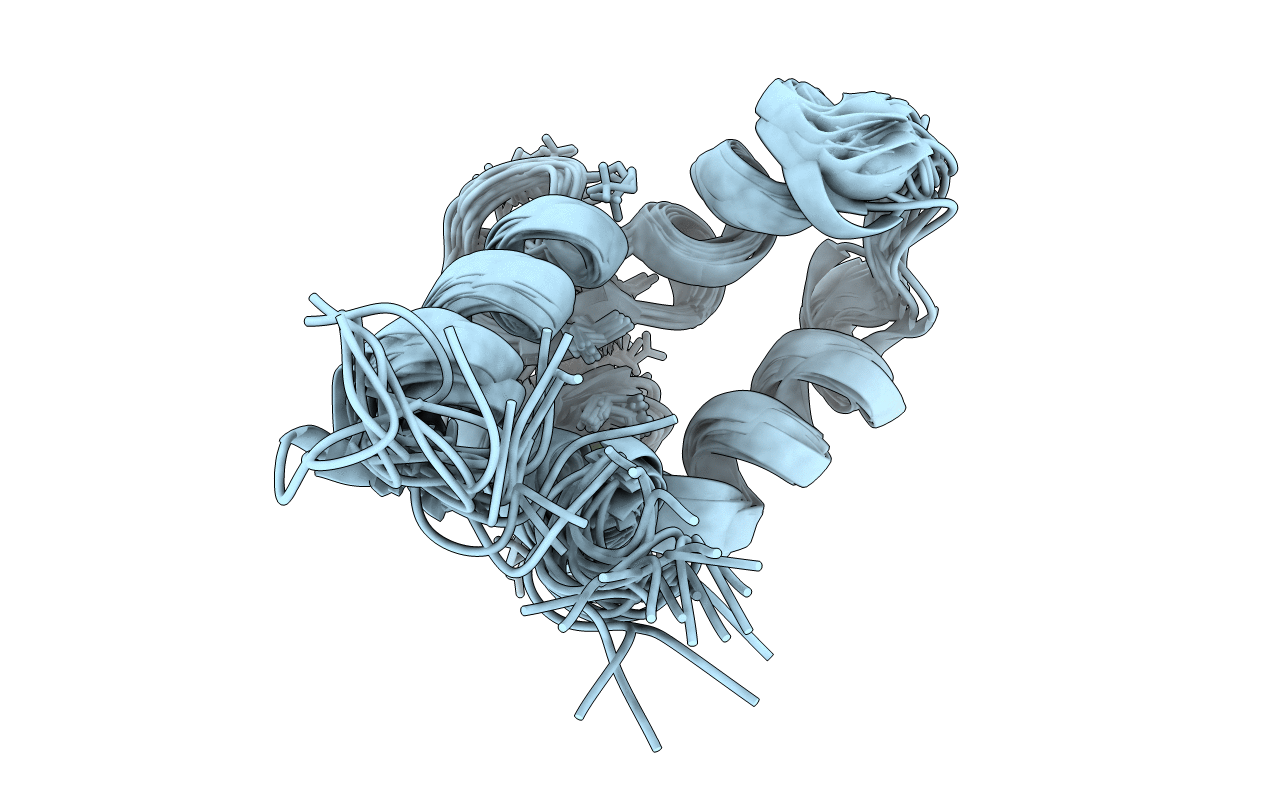
Deposition Date
2014-02-26
Release Date
2014-03-12
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2MLF
Keywords:
Title:
NMR structure of the dilated cardiomyopathy mutation G159D in troponin C bound to the anchoring region of troponin I
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
20
Selection Criteria:
target function