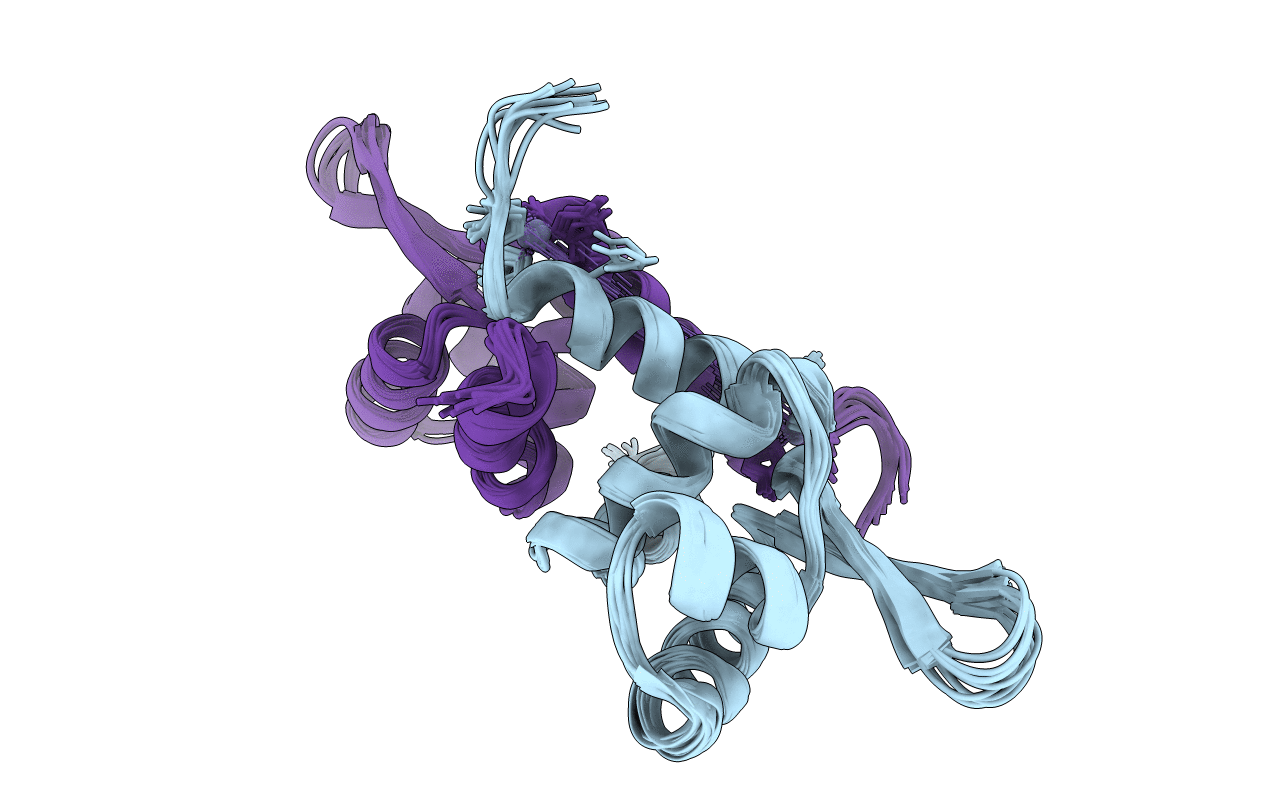
Deposition Date
2013-01-04
Release Date
2013-05-08
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2M30
Keywords:
Title:
Solution NMR refinement of a metal ion bound protein using quantum mechanical/molecular mechanical and molecular dynamics methods
Biological Source:
Source Organism(s):
Staphylococcus aureus (Taxon ID: 1280)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
132000
Conformers Submitted:
10
Selection Criteria:
Structures from 1 ns of independent QM/MM MD sampling


