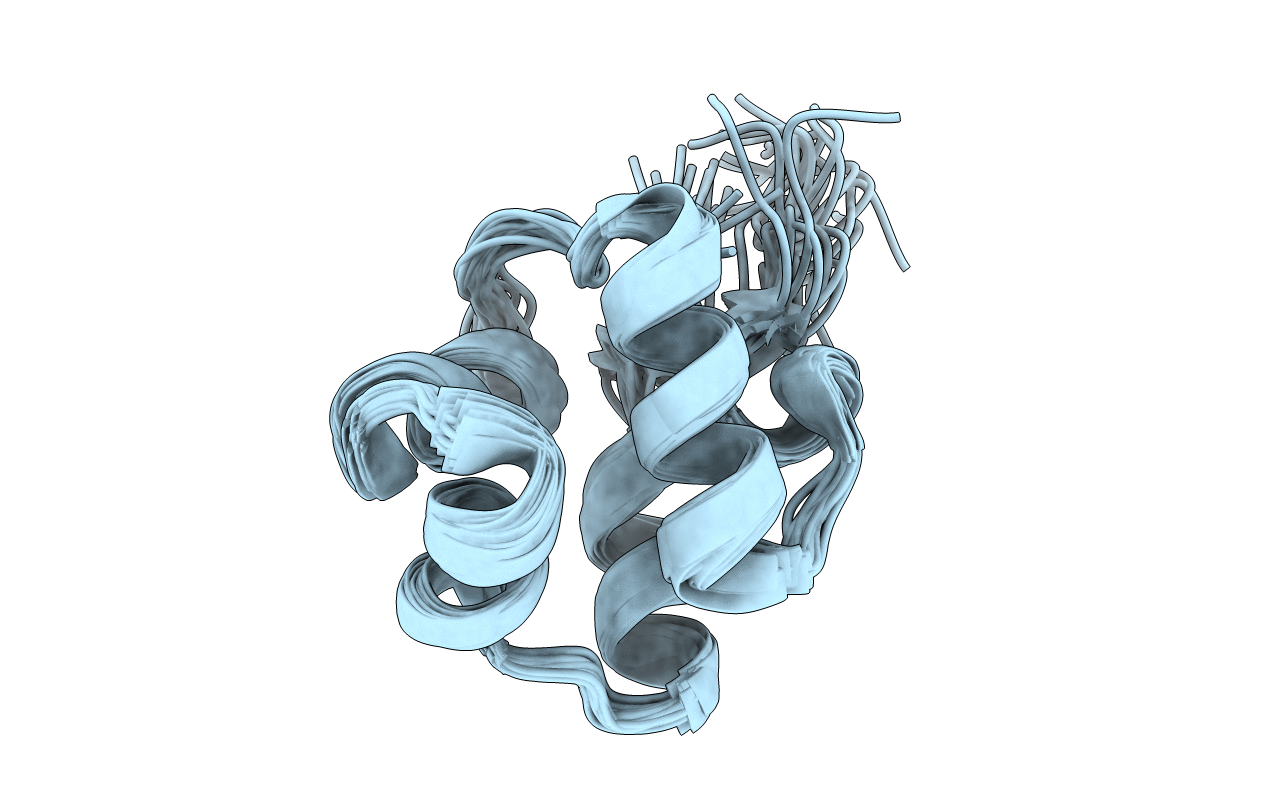
Deposition Date
2012-09-19
Release Date
2013-02-20
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2LYP
Keywords:
Title:
NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees)
Biological Source:
Source Organism(s):
Enterococcus faecalis (Taxon ID: 1351)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy