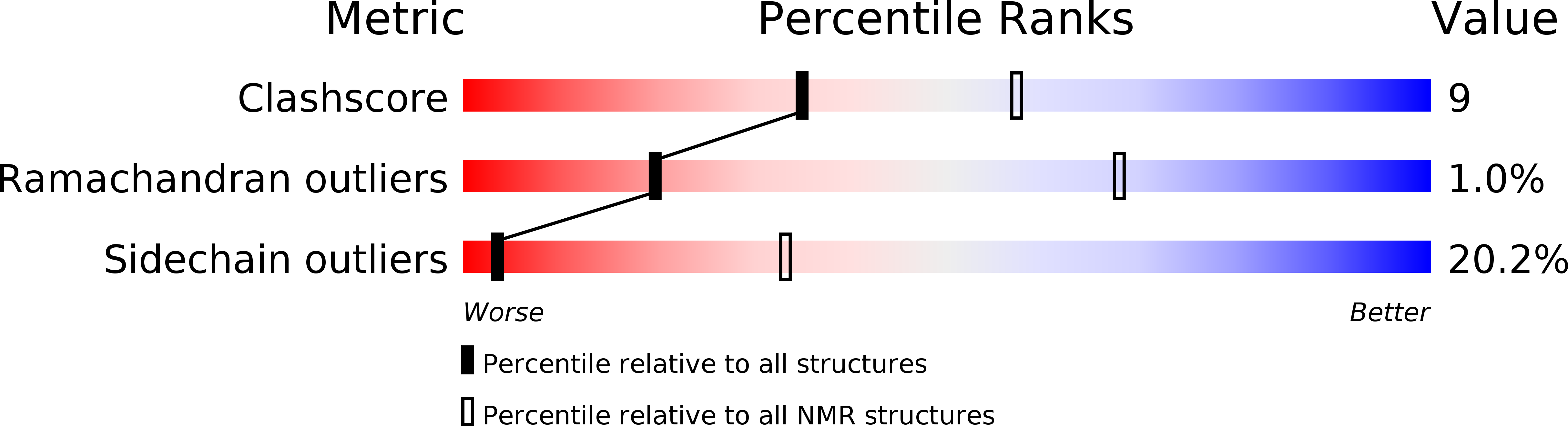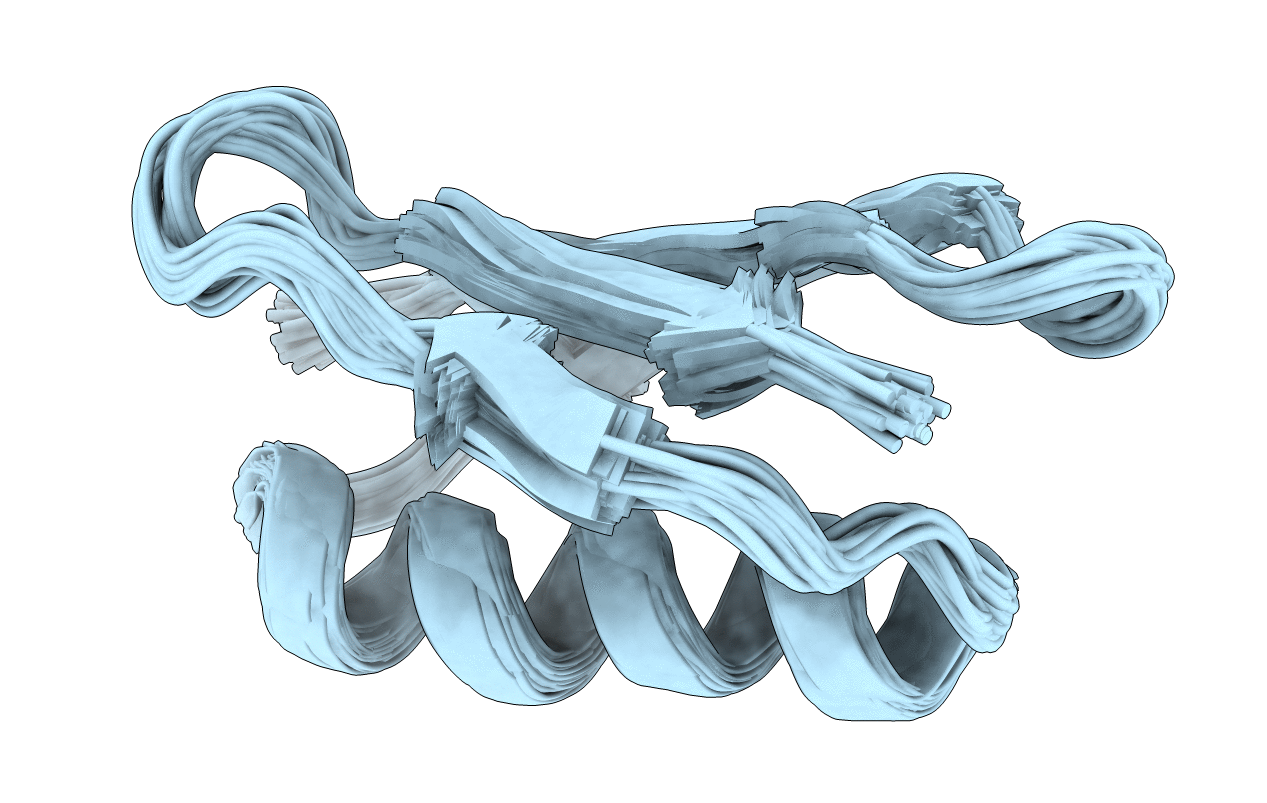
Deposition Date
2012-06-18
Release Date
2012-08-29
Last Version Date
2024-05-15
Entry Detail
PDB ID:
2LUM
Keywords:
Title:
Three-State Ensemble obtained from eNOEs of the Third Immunoglobulin Binding Domain of Protein G (GB3)
Biological Source:
Source Organism(s):
Streptococcus sp. 'group G' (Taxon ID: 1320)
Expression System(s):
Method Details: