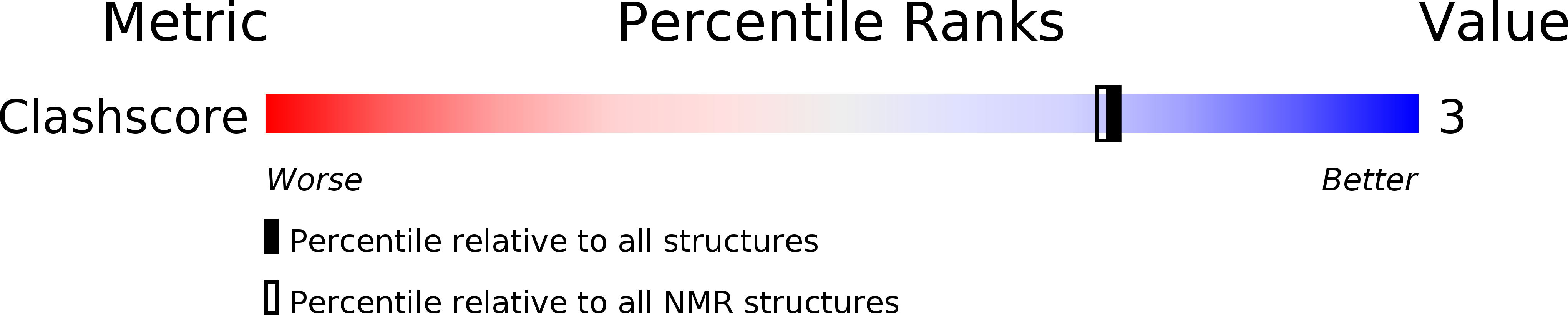Deposition Date
2012-04-30
Release Date
2012-09-12
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2LSF
Keywords:
Title:
Structure and Stability of Duplex DNA Containing (5'S) 5',8-Cyclo-2'-Deoxyadenosine: An Oxidative Lesion Repair by NER
Method Details:
Experimental Method:
Conformers Calculated:
26
Conformers Submitted:
26
Selection Criteria:
all calculated structures submitted