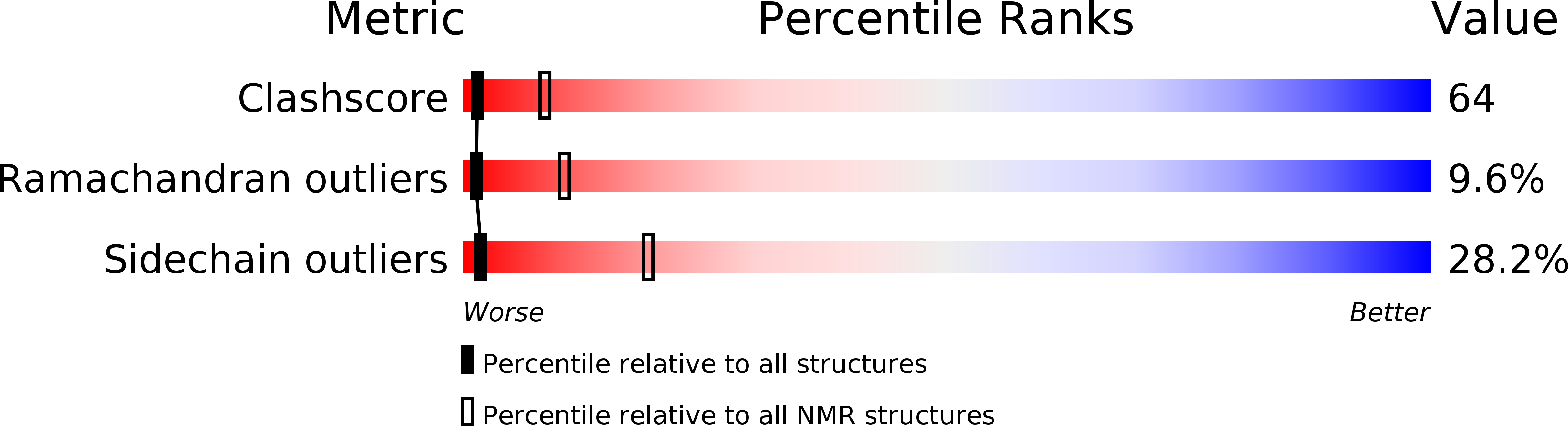
Deposition Date
2012-04-20
Release Date
2013-05-01
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2LS6
Keywords:
Title:
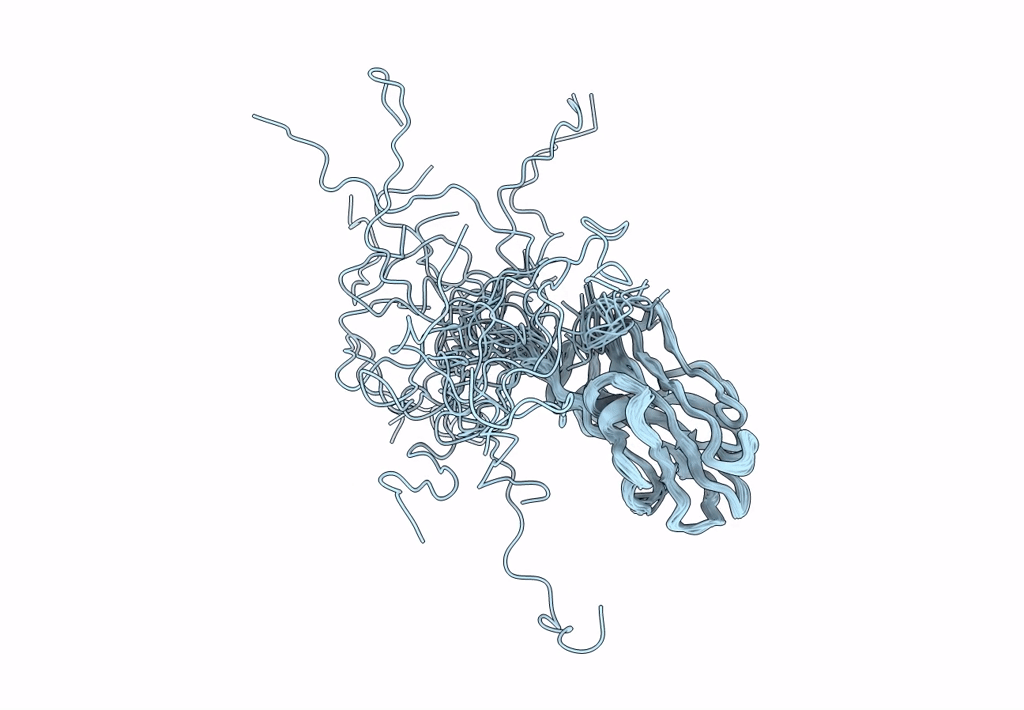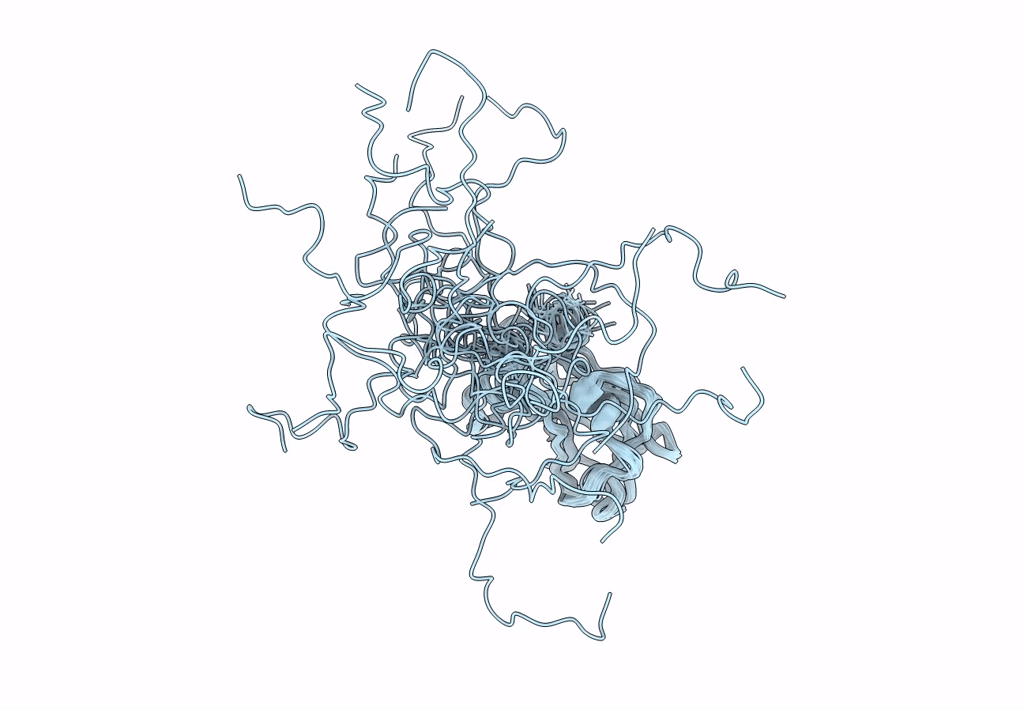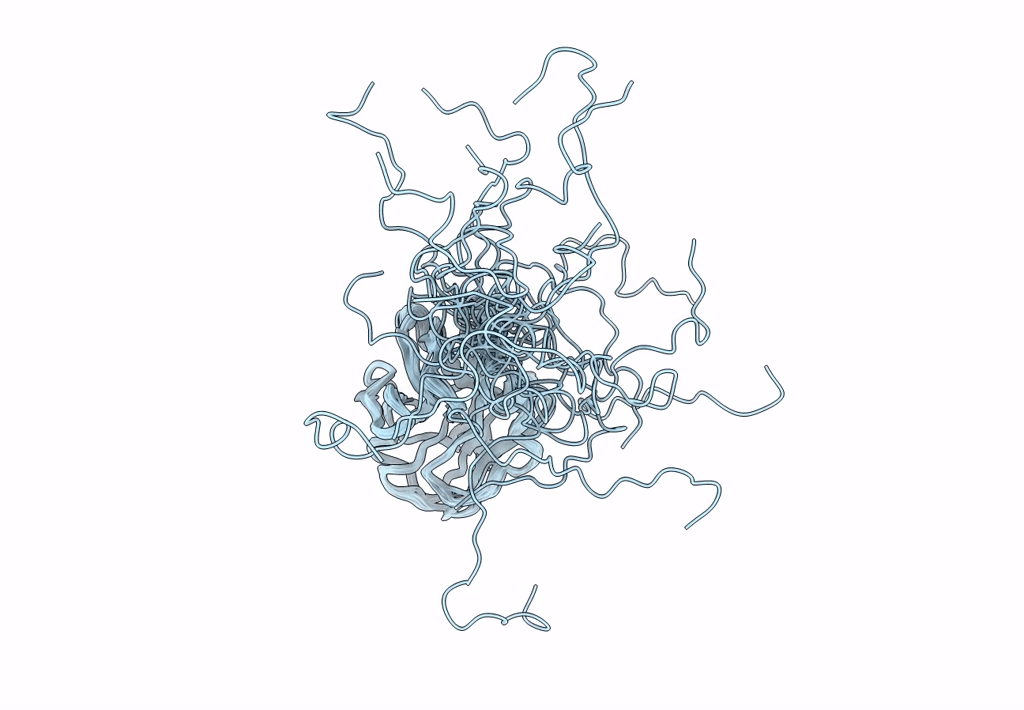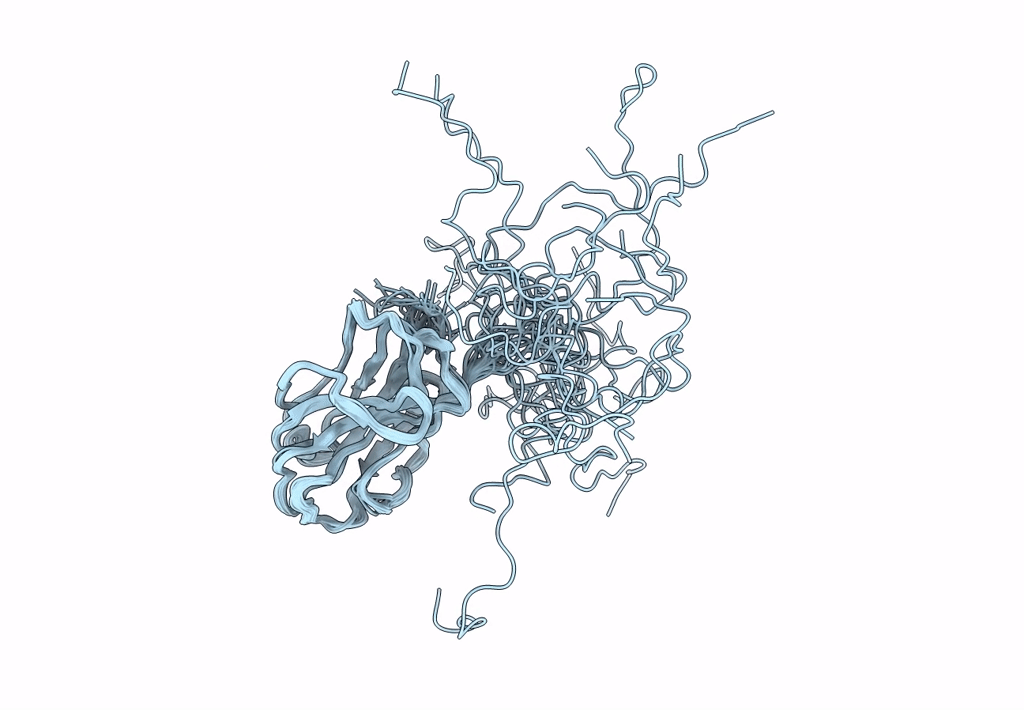
Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringens
Biological Source:
Source Organism(s):
Clostridium perfringens (Taxon ID: 195103)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
21
Selection Criteria:
structures with the lowest energy