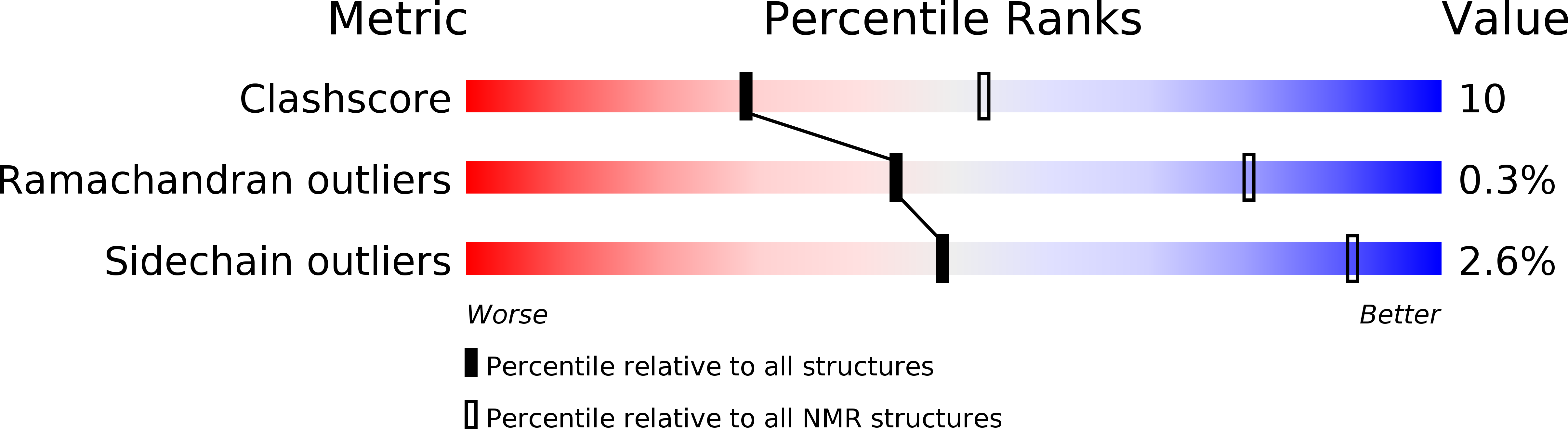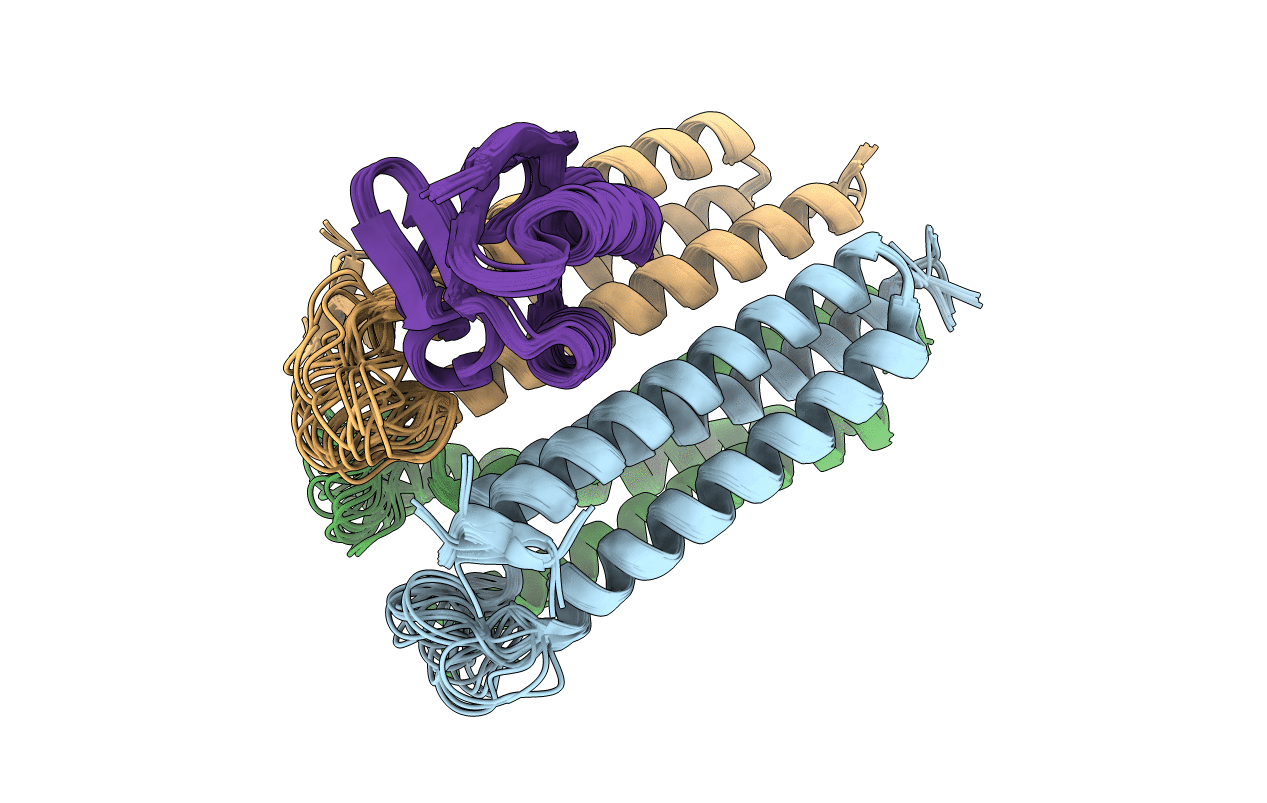
Deposition Date
2012-04-06
Release Date
2012-05-16
Last Version Date
2024-05-01
Entry Detail
PDB ID:
2LRK
Keywords:
Title:
Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
21
Selection Criteria:
structures with the least restraint violations